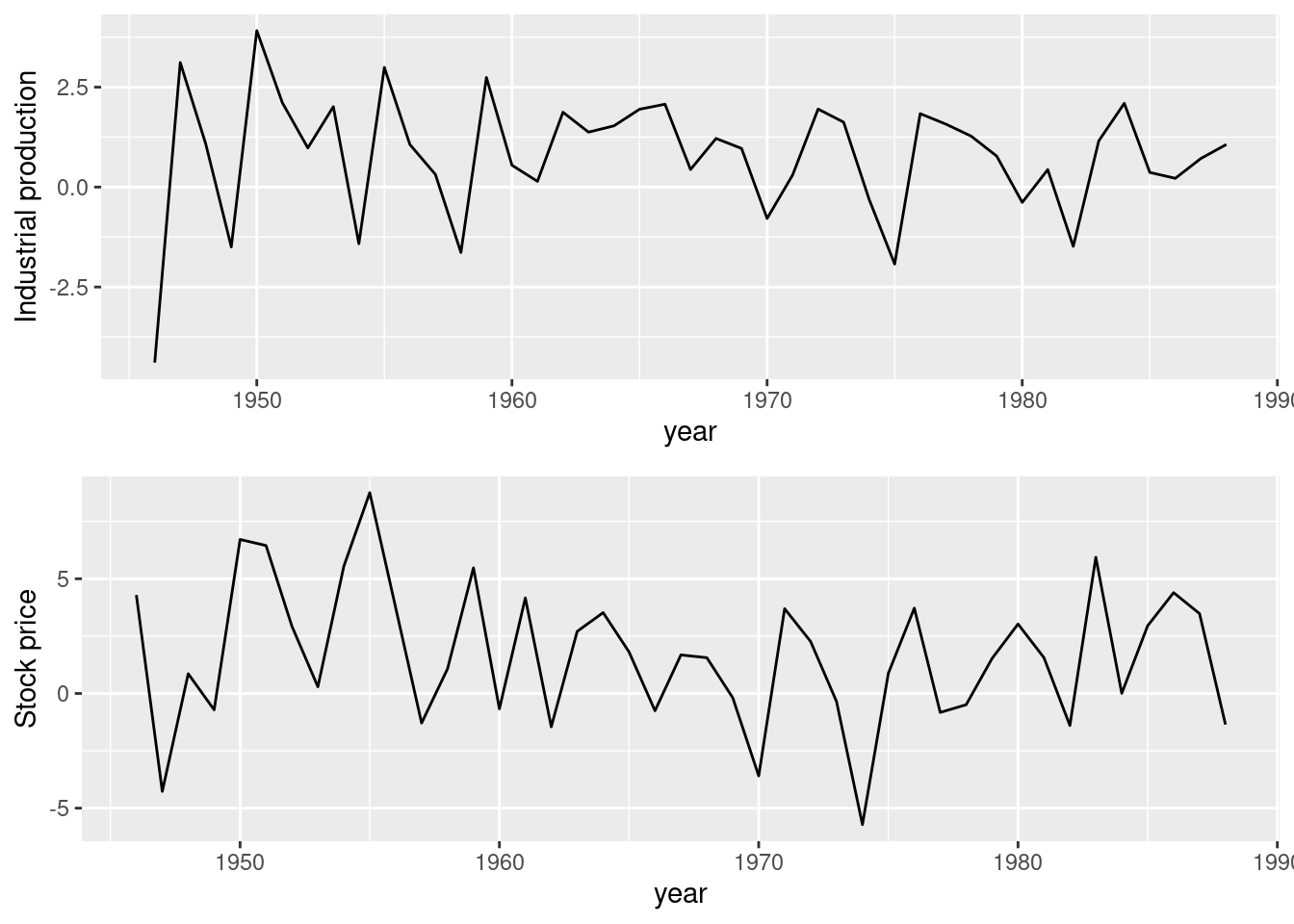
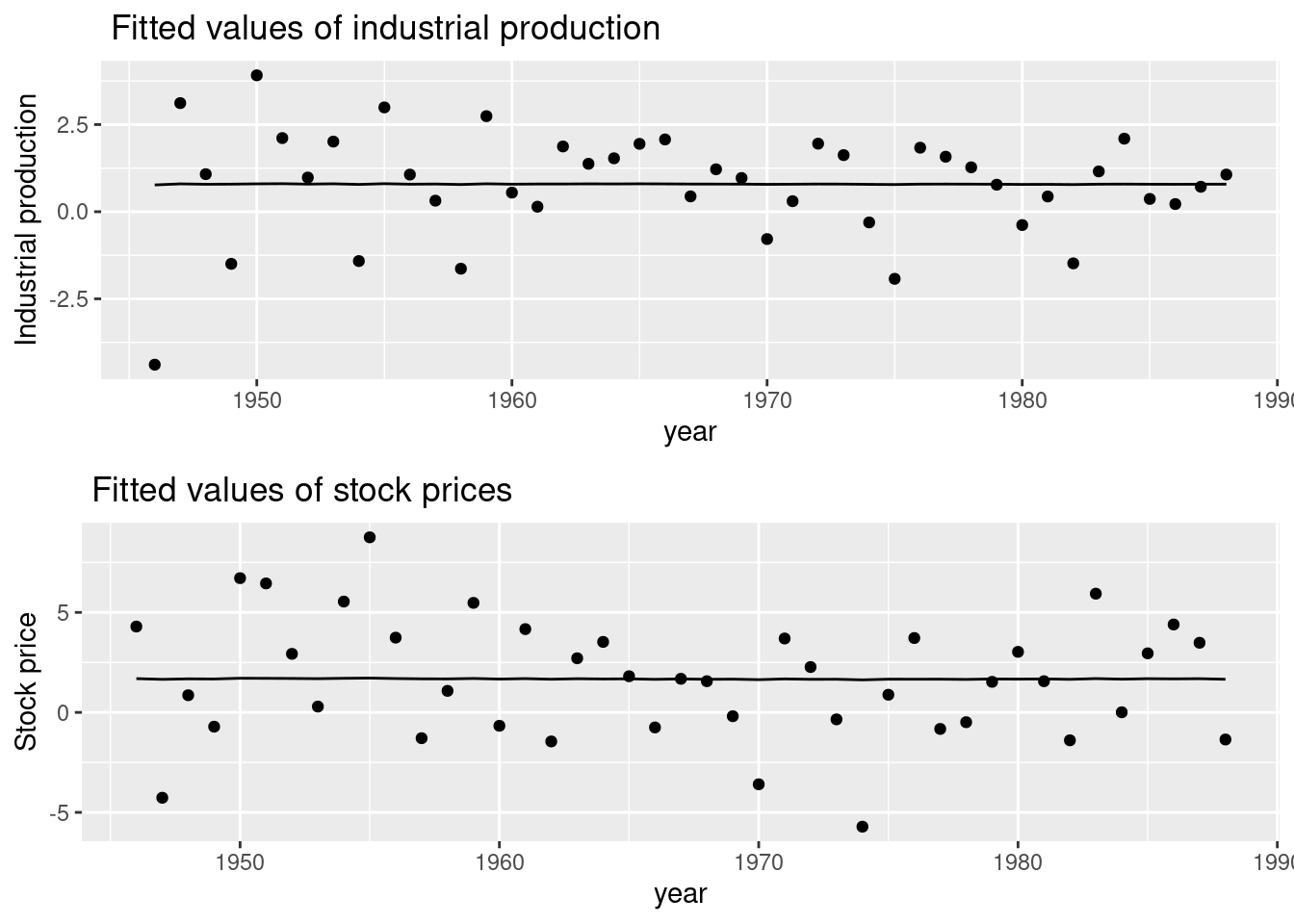
Chapter 6 Advanced Features
6.1 Introduction
As seen in previous chapters, INLA is a methodology to fit Bayesian hierarchical
models by computing approximations of the posterior marginal distributions of
the model parameters. In order to build more complex models and compute the
posterior marginal distribution of some quantities of interest, the INLA
package has a number of advanced features that are worth knowing. Some of
these features are also described in detail in Krainski et al. (2018).
In this chapter we will describe some of these features. In particular, Section
6.2 shows how to define a predictor matrix that multiplies
all terms in the linear predictor, so that the actual linear predictor of the
model is a linear combination of the effects in the model formula. Next,
Section 6.3 presents how to define linear combinations of the
latent effects to compute their posterior distribution. Section
6.4 introduces the use of models with several likelihoods.
Models with several likelihoods that share some terms are in Section
6.5, where the copy and replicate features are described
in detail. These features are very useful when working with, for example, joint
models (see Chapter 10).
6.2 Predictor Matrix
The formula passed to the inla() function defines the model to be fit by
INLA, i.e., the formula defines the terms in the linear predictor.
However, sometimes we need to modify the model so that linear combinations
of these terms are used instead of simply the ones set in the formula.
INLA allows this by defining a matrix A, which is called predictor matrix so that the actual linear
predictor \(\bm\eta^*\) used is
\[ \bm\eta^* = A \bm\eta \]
Here, \(\bm\eta\) is the linear predictor defined in the formula. By default, this matrix is the identity matrix.
Matrix A that defines the linear combinations to be used is passed using
element A in the list passed to argument control.predictor (see Section
2.5 for details). Matrix A can be a dense matrix or a sparse one
of the types implemented in the Matrix package (Bates and Maechler 2019). The dimension
of the A matrix must be \(n\times n\), with \(n\) the number of observations in
the model.
In order to provide a simple example we will consider the cement dataset
described in Section 2.3. A very simple use of the A matrix is
to re-scale the linear predictor used in the model (see Krainski et al. 2018 for
a similar example), for example, to have it multiplied by a constant. In this
case, we will take the A matrix as a diagonal matrix with values equal to 5.
This means that the linear predictors, as defined by the formula, will be
multiplied by 5, and that the estimates of the terms in the linear predictor
will shrink in a similar way so that the actual linear predictor \(A \bm\eta\)
remains the same as in the example in Section 2.3. Note that the
fitted values will also remain the same.
library("MASS")
library("Matrix")
data(cement)
A <- Diagonal(n = nrow(cement), x = 5)
summary(A)## 13 x 13 diagonal Matrix of class "ddiMatrix"
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 5 5 5 5 5 5Note how in the code above matrix A is a sparse matrix. Hence, the model
using this predictor matrix can be fit:
m1.A <- inla(y ~ x1 + x2 + x3 + x4, data = cement,
control.predictor = list(A = A))
summary(m1.A)##
## Call:
## "inla(formula = y ~ x1 + x2 + x3 + x4, data = cement,
## control.predictor = list(A = A))"
## Time used:
## Pre = 0.422, Running = 0.0601, Post = 0.0262, Total = 0.508
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 12.482 13.997 -15.556 12.481 40.495 12.482 0
## x1 0.310 0.149 0.012 0.310 0.608 0.310 0
## x2 0.102 0.145 -0.188 0.102 0.391 0.102 0
## x3 0.020 0.151 -0.282 0.020 0.322 0.020 0
## x4 -0.029 0.142 -0.313 -0.029 0.255 -0.029 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 0.209 0.093 0.068 0.195
## 0.975quant mode
## Precision for the Gaussian observations 0.428 0.167
##
## Expected number of effective parameters(stdev): 5.00(0.00)
## Number of equivalent replicates : 2.60
##
## Marginal log-Likelihood: -67.60Note how the estimates of the fixed effects are divided by 5 as compared
to the estimates obtained with the model fit in Section 2.3.
This is because of the effect of the A matrix.
In general, the predictor matrix can be useful for non-trivial examples. For example, the predictor matrix plays an important role in the development of spatial models based on stochastic partial differential equations (Krainski et al. 2018). See Chapter 7 for details.
6.3 Linear combinations
The use of the predictor matrix introduced in the previous section allows us to define models where the actual linear predictors are linear combinations of the effects in the model formula. However, in some situations we are only interested in computing linear combinations on some effects without altering model fitting.
With INLA, linear combinations on the different latent effects can be defined
and their posterior marginals estimated. Note that these linear combinations do
not alter the fit model (as the use of a predictor matrix did). Linear
combinations are passed using argument lincomb to the inla() function.
Furthermore, argument control.lincomb can be used to set the parameters that
control how posterior marginals of linear combinations are computed. Table
6.1 shows the options available to control
how the linear combinations are fit.
| Argument | Default | Description |
|---|---|---|
precision |
1e09 |
Tiny noise added to compute approximation |
verbose |
TRUE |
Use verbose if verbose is enabled globally. |
Linear combinations are defined by setting the effects and the associated
coefficients that they have to make the linear combination. Two functions are
available to define linear combinations: inla.make.lincomb(), to define a
single linear combination, and inla.make.lincombs(), to define several linear
combinations at once.
Function inla.make.lincomb() takes named arguments with the values of the
coefficients of the linear combination. The names are the names of the effects
to be used in the linear combination. For fixed effects, values must be a single number, while for random effects it is a vector of coefficients of the same
length as the index of the random effect. For those values of the random
effect not in the linear combination, NA must be used.
For example, in the analysis of the cement data above we might be interested
in computing the difference between coefficients of covariates x1 and
x2. This could be defined as follows:
inla.make.lincomb(x1 = 1, x2 = -1)If the linear combination involves a random effect index from 1 to \(p\),
the linear combination can be defined by including a term with the name
of the latent random effect. The value now is not a single number, but
a vector of length \(p\) with the coefficients of the different elements
of the random effects. Those elements that do not appear in the
linear combination must have a value of NA.
For example, if a random effect named u of length 4 is included in a
model and we want to compute the difference between its two terms, this
can be set up as:
inla.make.lincomb(u = c(1, -1, NA, NA))Function inla.make.lincombs() can be used to set more than one linear
combination at a time. Instead of using a single value for the linear
combinations on the fixed effects, a vector of coefficients can be passed.
For example, say that we are interested in comparing the coefficient of
variable x1 to the other three coefficients. In the design of experiments
literature these types of comparisons are often referred to as contrasts.
This can be set as several differences using linear combinations as follows:
inla.make.lincomb(
x1 = c( 1, 1, 1),
x2 = c(-1, 0, 0),
x3 = c( 0, -1, 0),
x4 = c( 0, 0, -1)
)Once the model has been fit, summaries of the linear combinations are available
in summary.lincomb.derived in the inla object returned. Similarly,
posterior marginals densities are available in marginals.lincomb.
Finally, two arguments can be passed through control.inla to control how
linear combinations are computed: lincomb.derived.only and
lincomb.derived.correlation.matrix. The first option indicates whether the
linear combinations are to be computed, and the second controls the computation
of the correlation matrix of the linear combinations. See Table
6.2 for a summary.
| Argument | Default | Description |
|---|---|---|
lincomb.derived.only |
TRUE |
Compute full graph of derived linear combinations. |
lincomb.derived.correlation.matrix |
FALSE |
Compute correlations for derived linear combinations. |
Linear combinations can be used for a number of issues, such as computing the posterior marginal of the linear predictor for a given set of covariate values. Another interesting application is to compare whether two effects are equal.
In order to provide a simple example, we will consider the abrasion dataset
(Davies 1954) from package faraway (Faraway 2016). This dataset records
data from an industrial experiment about wear of four different types of
materials after being fed into a wear testing machine. In each run,
four samples are processed, and the position of the samples may be important.
The different variables in this dataset are described in Table 6.3.
| Variable | Description |
|---|---|
run |
Run number (from 1 to 4). |
position |
Position number (from 1 to 4). |
material |
Type of material (A, B, C or D). |
wear |
Weight loss (in 0.1mm) over testing period. |
Given that the main interest is in the wear depending on the type of material,
material will be introduced into the model as a fixed effect, while run and
position are introduced as iid random affects. In addition, we have
noticed that default INLA priors produce too much shrinkage of the
coefficients and random effects towards zero, and the results do not reproduce
those in Faraway (2006). In particular, we have considered a larger precision on
the coefficients of the fixed effects. For the
prior on the precisions we have followed J. J. Faraway (2019b) and taken a Gamma prior
with parameters 0.5 and 95. This is an informative prior that has a mean
value half the variance of the residuals of a linear regression on wear
using material as predictor. The posterior means of precisions of the random
effects should be lower than this value. Furthermore, function
inla.hyperpar() has been used to obtain better estimates of the
hyperparameters once the model has been fit with INLA.
library("faraway")
data(abrasion)
# Prior prec. of random effects
prec.prior <- list(prec = list(param = c(0.5, 95)))
# Model formula
f.wear <- wear ~ -1 + material +
f(position, model = "iid", hyper = prec.prior) +
f(run, model = "iid", hyper = prec.prior)
# Model fitting
m0 <- inla(f.wear, data = abrasion,
control.fixed = list(prec = 0.001^2)
)
# Improve estimates of hyperparameters
m0 <- inla.hyperpar(m0)
summary(m0)##
## Call:
## "inla(formula = f.wear, data = abrasion, control.fixed = list(prec
## = 0.001^2))"
## Time used:
## Pre = 0.241, Running = 1.64, Post = 0.0965, Total = 1.98
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## materialA 265.7 9.784 246.0 265.7 285.3 265.7 0
## materialB 219.9 9.784 200.3 219.9 239.5 219.9 0
## materialC 241.7 9.784 222.0 241.7 261.3 241.7 0
## materialD 230.4 9.784 210.8 230.4 250.0 230.4 0
##
## Random effects:
## Name Model
## position IID model
## run IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 0.022 0.011 0.006 0.020
## Precision for position 0.008 0.006 0.001 0.006
## Precision for run 0.010 0.008 0.001 0.008
## 0.975quant mode
## Precision for the Gaussian observations 0.048 0.016
## Precision for position 0.025 0.004
## Precision for run 0.030 0.004
##
## Expected number of effective parameters(stdev): 9.28(0.43)
## Number of equivalent replicates : 1.72
##
## Marginal log-Likelihood: -98.70Then, we may be interested in comparing whether there is any difference between material A and any of the others. For example, the following linear constraint will compute the difference of the effects between material A and B:
lc <- inla.make.lincomb(materialA = 1, materialB = -1)This model can now be fit to compare materials A and B:
m0.lc <- inla.hyperpar(inla(f.wear, data = abrasion, lincomb = lc,
control.fixed = list(prec = 0.001^2)))
summary(m0.lc)##
## Call:
## "inla(formula = f.wear, data = abrasion, lincomb = lc,
## control.fixed = list(prec = 0.001^2))"
## Time used:
## Pre = 0.258, Running = 1.98, Post = 0.124, Total = 2.36
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## materialA 265.7 9.784 246.0 265.7 285.3 265.7 0
## materialB 219.9 9.784 200.3 219.9 239.5 219.9 0
## materialC 241.7 9.784 222.0 241.7 261.3 241.7 0
## materialD 230.4 9.784 210.8 230.4 250.0 230.4 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## lc 1 45.75 5.455 34.78 45.75 56.73 45.75 0
##
## Random effects:
## Name Model
## position IID model
## run IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 0.022 0.011 0.006 0.020
## Precision for position 0.008 0.006 0.001 0.006
## Precision for run 0.010 0.008 0.001 0.008
## 0.975quant mode
## Precision for the Gaussian observations 0.048 0.016
## Precision for position 0.025 0.004
## Precision for run 0.030 0.004
##
## Expected number of effective parameters(stdev): 9.28(0.43)
## Number of equivalent replicates : 1.72
##
## Marginal log-Likelihood: -98.70Note that summary statistics of the posterior marginal of the linear combination can be accessed with
m0.lc$summary.lincomb.derived## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## lc 1 45.75 5.455 34.78 45.75 56.73 45.75 0This shows a positive posterior means and 95% credible interval that is above 0, which means that wear in material A is higher than in material B.
This can be extended to compare material A to all the other materials. This
can be represented by a \(3\times 4\) matrix to represent the 3 linear combinations required. Now inla.make.lincombs must be used in order to define
3 different linear combinations.
lcs <- inla.make.lincombs(
materialA = c( 1, 1, 1),
materialB = c(-1, 0, 0),
materialC = c( 0, -1, 0),
materialD = c( 0, 0, -1)
)The model is fit similarly to the previous example:
m0.lcs <- inla.hyperpar(inla(f.wear, data = abrasion, lincomb = lcs,
control.fixed = list(prec = 0.001^2)))
summary(m0.lcs)##
## Call:
## "inla(formula = f.wear, data = abrasion, lincomb = lcs,
## control.fixed = list(prec = 0.001^2))"
## Time used:
## Pre = 0.261, Running = 2.58, Post = 0.0931, Total = 2.94
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## materialA 265.7 9.784 246.0 265.7 285.3 265.7 0
## materialB 219.9 9.784 200.3 219.9 239.5 219.9 0
## materialC 241.7 9.784 222.0 241.7 261.3 241.7 0
## materialD 230.4 9.784 210.8 230.4 250.0 230.4 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## lc1 1 45.75 5.455 34.78 45.75 56.73 45.75 0
## lc2 2 24.00 5.455 13.03 24.00 34.98 24.00 0
## lc3 3 35.25 5.455 24.28 35.25 46.23 35.25 0
##
## Random effects:
## Name Model
## position IID model
## run IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 0.022 0.011 0.006 0.020
## Precision for position 0.008 0.006 0.001 0.006
## Precision for run 0.010 0.008 0.001 0.008
## 0.975quant mode
## Precision for the Gaussian observations 0.048 0.016
## Precision for position 0.025 0.004
## Precision for run 0.030 0.004
##
## Expected number of effective parameters(stdev): 9.28(0.43)
## Number of equivalent replicates : 1.72
##
## Marginal log-Likelihood: -98.70Linear combinations can also be used to compare the levels of the random
effects. For example, let’s consider a comparison between positions 1 and
2. Variable position is an iid random effect with four different levels,
and the weights must be set in a vector of the same length. For those
levels that must be ignored in the linear combination, NA can be used.
Hence, to define a linear combination to compare levels 1 and 2 of variable
position we can define the linear combination as follows:
lc.pos <- inla.make.lincomb(position = c(1, -1, NA, NA))
m0.pos <- inla.hyperpar(inla(f.wear, data = abrasion, lincomb = lc.pos,
control.fixed = list(prec = 0.001^2)))
summary(m0.pos)##
## Call:
## "inla(formula = f.wear, data = abrasion, lincomb = lc.pos,
## control.fixed = list(prec = 0.001^2))"
## Time used:
## Pre = 0.247, Running = 1.88, Post = 0.0957, Total = 2.22
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## materialA 265.7 9.784 246.0 265.7 285.3 265.7 0
## materialB 219.9 9.784 200.3 219.9 239.5 219.9 0
## materialC 241.7 9.784 222.0 241.7 261.3 241.7 0
## materialD 230.4 9.784 210.8 230.4 250.0 230.4 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## lc 1 -23.34 5.504 -33.67 -23.59 -11.62 -24.09 0
##
## Random effects:
## Name Model
## position IID model
## run IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 0.022 0.011 0.006 0.020
## Precision for position 0.008 0.006 0.001 0.006
## Precision for run 0.010 0.008 0.001 0.008
## 0.975quant mode
## Precision for the Gaussian observations 0.048 0.016
## Precision for position 0.025 0.004
## Precision for run 0.030 0.004
##
## Expected number of effective parameters(stdev): 9.28(0.43)
## Number of equivalent replicates : 1.72
##
## Marginal log-Likelihood: -98.70Fixed and random effects can be combined together to create more complex linear combinations. For example, the following linear combination considers the sum of the effect of material A, position 1 and run 2:
lc.eff <- inla.make.lincomb(materialA = 1, position = c(1, NA, NA, NA),
run = c(NA, 1, NA, NA))
m0.eff <- inla.hyperpar(inla(f.wear, data = abrasion, lincomb = lc.eff,
control.fixed = list(prec = 0.001^2)))
summary(m0.eff)##
## Call:
## "inla(formula = f.wear, data = abrasion, lincomb = lc.eff,
## control.fixed = list(prec = 0.001^2))"
## Time used:
## Pre = 0.248, Running = 1.82, Post = 0.101, Total = 2.17
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## materialA 265.7 9.784 246.0 265.7 285.3 265.7 0
## materialB 219.9 9.784 200.3 219.9 239.5 219.9 0
## materialC 241.7 9.784 222.0 241.7 261.3 241.7 0
## materialD 230.4 9.784 210.8 230.4 250.0 230.4 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## lc 1 254 5.906 242.6 253.9 266.3 253.6 0
##
## Random effects:
## Name Model
## position IID model
## run IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 0.022 0.011 0.006 0.020
## Precision for position 0.008 0.006 0.001 0.006
## Precision for run 0.010 0.008 0.001 0.008
## 0.975quant mode
## Precision for the Gaussian observations 0.048 0.016
## Precision for position 0.025 0.004
## Precision for run 0.030 0.004
##
## Expected number of effective parameters(stdev): 9.28(0.43)
## Number of equivalent replicates : 1.72
##
## Marginal log-Likelihood: -98.706.4 Several likelihoods
So far, we have considered models with a single likelihood but INLA can
handle models with more than one likelihood. This is useful to define
joint models (see Chapter 10) in which one likelihood
models survival time and the second one models longitudinal data.
The first likelihood will be that of a survival model, while the second
one will be of the types used to model longitudinal data.
When using several likelihoods data must be stored in a very particular way.
First of all, the response variable must be a matrix with as many columns as
likelihoods. Hence, the first column will be the response used by the first
likelihood and so on. Data must be stored so that there is one variable per
column and a single value of any of the variables per row. All the empty
elements in the matrix are set to NA.
For example, let us consider a joint model with response variables \(\mathbf{y} = (y_1\ldots, y_n)\) and \(\mathbf{z} = (z_1,\ldots, z_m)\). Then, data must be stored in a matrix like the following:
\[ \left[ \begin{array}{cc} y_1 & \mathtt{NA}\\ \vdots & \mathtt{NA}\\ y_n & \mathtt{NA}\\ \mathtt{NA} & z_1\\ \mathtt{NA} & \vdots\\ \mathtt{NA} & z_m\\ \end{array} \right] \]
Note that the dimension of this matrix is \((n + m) \times 2\), i.e., the number of rows is the total number of observations and the number of columns is equal to the number of variables. This can be easily generalized to any number of response variables.
When multiple likelihoods are used in a model, these are passed in a vector
to argument family. For example, the following code could be used to fit
a model with a Gaussian and a Poisson likelihood:
family = c("gaussian", "poisson")As stated above, the response variables need to be formatted in a particular way. A matrix with as many likelihoods as columns is required. The number of rows is the total number of observations for all likelihoods. If in the example above there are 30 observations from a Gaussian likelihood and 20 from a Poisson one, then the response must be a matrix with 2 columns and 50 rows.
Then, observations are stacked in a matrix so that the observations from the
Gaussian likelihood are in the first column and rows 1 to 30. Then, observations
for the Poisson likelihood are in the second column and rows 31 to 50. All the
other elements in the matrix are filled with NA. This ensures that every
row has only a single observed value. Note that then there is an implicit
index from 1 to 50 to identify each one of the observations, regardless of
the likelihood they belong to.
In order to define the model, all terms for both likelihoods need to be
included in the linear predictor. Hence, when defining indices for the latent
random effects, this can go (for example) from 1 to the total number of rows in
the response matrix. When a covariate only affects
observations in a single likelihood, the values for the observations in the
other likelihoods must be set to NA. Similarly, the indices of the latent
random effects must be set to NA for those observations that do not depend
on the latent random effect.
6.4.1 Simulated example
To illustrate the use of several likelihoods we will develop an example here using two (independent) variables from a Gaussian and Poisson likelihood, respectively. First of all, 30 observations from a standard Gaussian distribution and 20 observations from a Poisson with mean 10 will be drawn:
set.seed(314)
# Gaussian data
d1 <- rnorm(30)
# Poisson data
d2 <- rpois(20, 10)Next, the two response variables will be put in a 2-column matrix as
required by INLA:
# Data
d <- matrix(NA, ncol = 2, nrow = 30 + 20)
d[1:30, 1] <- d1
d[30 + 1:20, 2] <- d2In order to have two different intercepts in the model it is necessary to add
them as two separate covariates. In this case, both intercepts have all
values equal to one, but the same principle would apply when two separate
coefficients are required for the same covariate or when random effects only
apply to data in one the likelihoods. Values NA mean that the covariate does
not appear in the linear predictor of the response.
# Define a different intercept for each likelihood
Intercept1 <- c(rep(1, 30), rep(NA, 20))
Intercept2 <- c(rep(NA, 30), rep(1, 20))If the coefficient is to be the same, shared between both likelihoods, then the
covariate is replicated as many times as likelihoods in the model, so that it
is a vector. For example, the next variable x will be shared by both terms
because it is a vector of simulated data of length 50 (the total number of
observations):
x <- rnorm(30 + 20)Finally, the model is defined and fit below. Note how the data is now
passed using a list as a data.frame would not be suitable in this case
because the response is a matrix.
mult.lik <- inla(Y ~ -1 + I1 + I2 + x,
data = list(Y = d, I1 = Intercept1, I2 = Intercept2, x = x),
family = c("gaussian", "poisson"))
summary(mult.lik)##
## Call:
## c("inla(formula = Y ~ -1 + I1 + I2 + x, family = c(\"gaussian\",
## \"poisson\"), ", " data = list(Y = d, I1 = Intercept1, I2 =
## Intercept2, x = x))" )
## Time used:
## Pre = 0.197, Running = 0.0827, Post = 0.0171, Total = 0.297
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## I1 -0.316 0.161 -0.635 -0.316 0.003 -0.316 0
## I2 2.222 0.077 2.067 2.223 2.369 2.225 0
## x 0.021 0.071 -0.118 0.021 0.159 0.021 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 1.36 0.346 0.769 1.33
## 0.975quant mode
## Precision for the Gaussian observations 2.12 1.27
##
## Expected number of effective parameters(stdev): 3.00(0.00)
## Number of equivalent replicates : 16.66
##
## Marginal log-Likelihood: -117.94Note that the second intercept (for the Poisson likelihood) is close to the
logarithm of the actual mean and not the actual mean of the Poisson likelihood.
Also, there is a single coefficient for x because it is shared by both parts
of the model. The point estimate is very close to zero because the covariate
had no effect on the response when the data were simulated.
6.6 Linear constraints
As introduced in Chapter 3, additional linear constraints on the latent effects can be imposed. Given a latent effect \(\mathbf{u}\), the linear constraint is defined as
\[ \mathbf{A} \mathbf{u}^{\top} = \mathbf{e}^{\top} \]
where \(\mathbf{A}\) is a matrix with the same number of columns as the length of the latent effect \(\mathbf{u}\). Hence, the number of rows represents the number of linear constraints on the latent effect and \(\mathbf{e}\) is a vector of values that is defined accordingly.
Both \(\mathbf{A}\) and \(\mathbf{e}\) are passed through argument extraconstr
using a named list with values A and e in the the definition of the
latent effect using function f(). Note that there is another
argument constr which can be used to add a sum-to-zero constraint on
the latent effect by setting it to TRUE. This argument is set to
TRUE for some latent effects (such as, for example, the rw1
and besag latent effects and other intrinsic effects). In case of doubt,
the default option can be seen in the documentation available via the
inla.doc() function.
| Argument | Default | Description |
|---|---|---|
constr |
Depends on latent effect. | Add sum-to-zero constraint. |
extraconstr |
list(A = NULL, e = NULL) |
Add extra constraints on the random effect. |
Adding linear constraints is important in some cases as this makes the different effects in a model identifiable. Rue and Held (2005) describe this topic in detail. Furthermore, see Knorr-Held (2000) and Ugarte et al. (2014) on the role of imposing linear constraints to spatio-temporal latent effects, but these principles could also be applied to other types of latent effects. Also, Goicoa et al. (2018) note that adding additional constraints on the latent effects may produce different estimates than expected and caution should be taken when setting additional linear constraints.
As a simple example, we will add a sum to zero constraint to the ar1
model fit to the NelPLo dataset. In this case only the industrial production
will be considered. We will also remove the intercept to show the effect of
constraining the random effect.
First, the matrix A and vector e need to be defined:
# Define values of A and e to set linear constraints
A <- matrix(1, ncol = n, nrow = 1)
e <- matrix(0, ncol = 1)Next, the model is fit using argument extraconstr in the call to the
f() function that defines the latent effect. Remember that the intercept
has the sole purpose of making the effect of the sum-to-zero constraint
more evident.
m.unconstr <- inla(ip ~ -1 + f(idx, model = "ar1"),
data = list(ip = NelPlo[, 1], idx = 1:n),
control.family = list(hyper = list(prec = list(initial = 10,
fixed = TRUE))),
control.predictor = list(compute = TRUE)
)
m.constr <- inla(ip ~ -1 + f(idx, model = "ar1",
extraconstr = list(A = A, e = e)),
data = list(ip = NelPlo[, 1], idx = 1:n),
control.family = list(hyper = list(prec = list(initial = 10,
fixed = TRUE))),
control.predictor = list(compute = TRUE)
)Figure 6.3 shows the estimates of the random effects from both models. Note how now imposing the sum-to-zero constraint seems to provide slightly different estimates from the unconstrained estimates. It is possible that the estimates of other parameters in the model also change as a result of imposing a constraint on the random effects, as described in Goicoa et al. (2018).
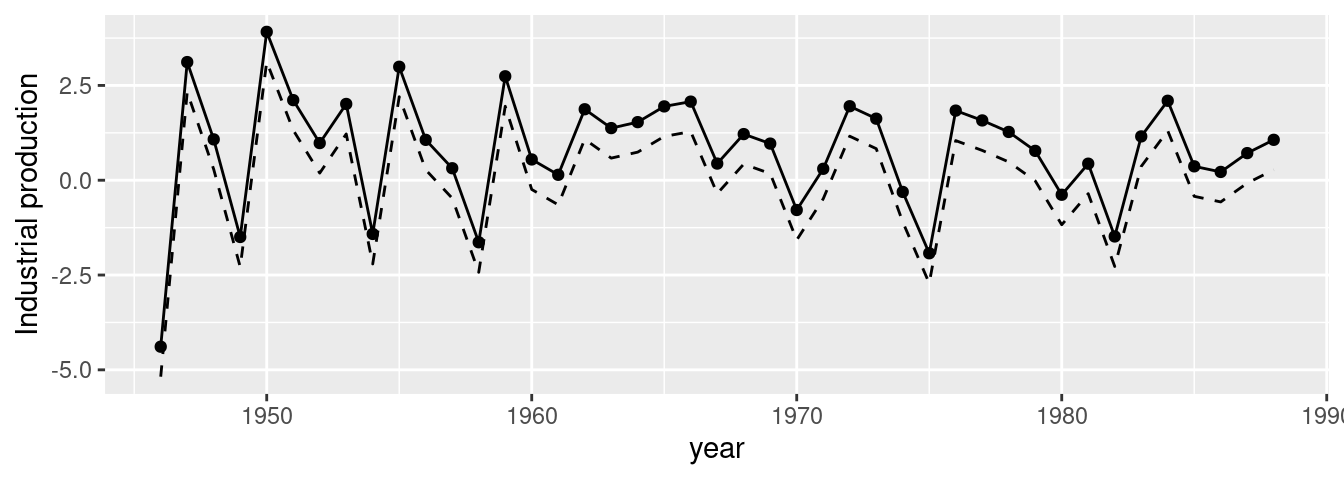
Figure 6.3: Comparison between two random effects estimates using a sum-to-zero constraint (solid line) and no constraint (dashed lines).
6.7 Final remarks
The features presented in this chapter can help to build more complex models
using INLA. These are extensively used in some of the models used in other
parts of the book. When the latent effect has a very complex structure
that does not fit any of the latent effects described so far with the
help of these additional features, there are a number of ways to implement
custom latent effects for INLA. These methods are described in
Chapter 11.
References
Bates, Douglas, and Martin Maechler. 2019. Matrix: Sparse and Dense Matrix Classes and Methods. https://CRAN.R-project.org/package=Matrix.
Davies, O. L. 1954. The Design and Analysis of Industrial Experiments. Wiley.
Faraway, J. 2006. Extending the Linear Model with R. Chapman; Hall, London.
Faraway, Julian. 2016. Faraway: Functions and Datasets for Books by Julian Faraway. https://CRAN.R-project.org/package=faraway.
Faraway, Julian J. 2019b. “INLA for Linear Mixed Models.” http://www.maths.bath.ac.uk/~jjf23/inla/.
Goicoa, T., A. Adin, M. D. Ugarte, and J. S. Hodges. 2018. “In Spatio-Temporal Disease Mapping Models, Identifiability Constraints Affect PQL and INLA Results.” Stochastic Environmental Research and Risk Assessment 3 (32): 749–70.
Knorr-Held, Leonhard. 2000. “Bayesian Modelling of Inseparable Space-Time Variation in Disease Risk.” Statistics in Medicine 19 (17‐18): 2555–67.
Koop, G., and M. F. J. Steel. 1994. “A Decision-Theoretic Analysis of the Unit-Root Hypothesis Using Mixtures of Elliptical Models.” Journal of Business and Economic Statistics 12: 95–107.
Krainski, Elias T., Virgilio Gómez-Rubio, Haakon Bakka, Amanda Lenzi, Daniela Castro-Camilo, Daniel Simpson, Finn Lindgren, and Håvard Rue. 2018. Advanced Spatial Modeling with Stochastic Partial Differential Equations Using R and INLA. Boca Raton, FL: Chapman & Hall/CRC.
Petris, Giovanni. 2010. “An R Package for Dynamic Linear Models.” Journal of Statistical Software 36 (12): 1–16. http://www.jstatsoft.org/v36/i12/.
Petris, Giovanni, Sonia Petrone, and Patrizia Campagnoli. 2009. Dynamic Linear Models with R. UseR! Springer-Verlag, New York.
Rue, H., and L. Held. 2005. Gaussian Markov Random Fields: Theory and Applications. Chapman; Hall/CRC Press.
Ugarte, M. D., A. Adin, T. Goicoa, and A. F. Militino. 2014. “On Fitting Spatio-Temporal Disease Mapping Models Using Approximate Bayesian Inference.” Statistical Methods in Medical Research 23: 507–30. https://doi.org/10.1177/0962280214527528.