Chapter 10 Survival Models
10.1 Introduction
Survival data analysis tackles the problem of modeling observations of time to
event. In this context, the interest is the time until a certain event
happens. This can be death (e.g., survival time since diagnosis) or failure
(e.g., time until a piece breaks down). We will give a brief overview of
survival analysis now, in order to introduce the main concepts and define
our notation. A more comprehensive treatment of Bayesian survival analysis can be found in Ibrahim, Chen, and Sinha (2001). Moore (2016) also provides a nice introduction to survival analysis with R.
Considering T as the random variable that measures time to event, the survival function \(S(t)\) can be defined as the probability that \(T\) is higher than a given time \(t\), i.e., \(S(t) = P(T > t)\). For example, this could be the probability that a patient will survive at least until time \(t\) since diagnosis. In survival analysis there are the implicit assumptions that \(S(0) = 1\), i.e., all patients are alive at the beginning of the study, and that \(\lim_{t\to+\infty} S(t) = 0\), i.e., all patients will eventually die (Valar Morghullis).
When considering a survival function, it will be related to a distribution function \(F(t) = P(T <t) = 1 - S(t)\) with density function \(f(t)\). This can be a parametric distribution, as we will see below.
The hazard function \(h(t)\) measures the probability that the event occurs at a small instant given that the patient has survived until time \(t\). More precisely,
\[ h(t) = \lim_{\Delta t\to 0} \frac{P(T < t + \Delta t \mid T > t)}{\Delta t} \]
The cumulative hazard \(H(t)\) is defined as
\[ H(t) = \int_{0} ^t h(u) du \]
Hence, the hazard function can be expressed as \(h(t) = f(t) / S(t)\). Furthermore, the survival function can be expressed in terms of the cumulative hazard function as
\[ S(t) = \exp (-H(t)) \]
Survival data are often censored. For example, patients may experience the event during the study period, in which case the time to event is observed, or after the study has been finished, in which case the event time is unobserved. For this reason, each observation is accompanied by a status variable \(\delta_i\) to indicate whether the event was observed or not. In addition, a vector of covariates may be available for each patient. Hence, the data will be made of \(\{(t_i, \delta_i, \mathbf{x}_i\}_{i=1}^n\), where \(t_i\) is time of patient \(i\), \(\delta_i\) the associated censoring status variable and \(\mathbf{x}_i\) the covariates associated to patient \(i\).
The likelihood of this model can be expressed as:
\[ \prod_{i=1}^n f(t_i)^{\delta_i} S(t_i)^{1-\delta_i} \]
Note how depending on whether an observation has been censored its contribution to the likelihood will be either \(f(t_i)\) or \(S(t_i)\).
10.2 Non-parametric estimation of the survival curve
The Kaplan-Meier estimator provides a non-parametric estimate of the survival curve. It is based on the conditional probability of surviving until time \(t\) given that the patient has survived until time \(t_i\) and it is defined as
\[ \hat{S}(t) = \prod _{t_i \leq t} (1 - \frac{d_i}{n_i}) \]
Here, \(n_i\) is the number of subjects at risk at time \(i\) and \(d_i\) the number of individuals that fail at that time. Note that at a given time \(t_i\), \((1 - d_i/n_i)\) is the proportion of individuals at risk at time \(t_i\) that do not experience any event. Hence, the Kaplan-Meier estimate is the product of all conditional probabilities of not experiencing the event up to time \(t\).
The survival package (Terry M. Therneau and Patricia M. Grambsch 2000; Therneau 2015) implements a number
of functions for the analysis of survival data, including the Kaplan-Meier
estimator, and includes various datasets. We will be using the veteran
dataset (Kalbfleisch and Prentice 1980), which contains data from the Veteran’s
Administration Lung Cancer Trial. This dataset records time to death of 137
male patients with advanced inoperable lung cancer. Patients were assigned at
random a standard treatment or a test treatment. Only 9 patients left the trial
before they experienced death. In addition to the time to death and the
treatment received, there are other covariates measured and they have been
summarized in Table 10.1.
| Variable | Description |
|---|---|
trt |
Treatment (1 for standard, 2 for test treatment) |
celltype |
Histological type of tumor (1=squamous, 2=smallcell, 3=adeno, 4=large). |
time |
Survival time (in days). |
status |
Censoring status (1 for death, 0 for censored). |
karno |
Karnofsky performance score (100 means good). |
diagtime |
Months from diagnosis to randomization. |
age |
Age in years. |
prior |
Prior therapy (0 for no, 10 for prior treatment). |
First of all, we will load the data and recode some of the variables in the study as factors:
library("survival")
data(veteran)
veteran$trt <- as.factor(veteran$trt)
levels(veteran$trt) <- c("standard", "test")
veteran$prior <- as.factor(veteran$prior)
levels(veteran$prior) <- c("No", "Yes")As stated in the INLA documentation,
some likelihoods for survival data may be affected
by numerical overflow when observed times are large.
For this reason, we will re-scale time before fitting
any model and express time as 30-day months.
veteran$time.m <- round(veteran$time / 30, 3)We will use the
Surv() function to create a data structure with the required format for
data analysis using the survival/censoring times and the censoring indicator.
This is illustrated in the next lines where the first 20 values in the survival data structure are shown:
surv.vet <- Surv(veteran$time.m, veteran$status)
surv.vet[1:20]## [1] 2.400 13.700 7.600 4.200 3.933 0.333 2.733 3.667
## [9] 10.467 3.333+ 1.400 0.267 4.800 0.833+ 0.367 1.000
## [17] 12.800 0.133 1.800 0.433Note how censored observations are marked with a + to indicate
that the time to event has been censored and it is larger than the value shown.
The Kaplan-Meier estimator can be used to obtain an estimate of the survival function as follows:
km.vet <- survfit(surv.vet ~ 1)Figure 10.2 (left plot) shows a Kaplan-Meier estimate of the
survival time for the veteran dataset. The plot includes confidence intervals
as well as point estimates of the survival time. Note that the plot on
the right corresponds to a parametric estimate discussed later in
Section 10.3. They have been put side by side so that
the different estimates can be compared.
Furthermore, four different survival curves can be estimated by splitting the data according to the different types of tumors:
km.vet.cell <- survfit(surv.vet ~ -1 + celltype, data = veteran)Figure 10.1 shows estimates of the survival times for each of
the four groups of patients defined by the covariate. Here, function
autoplot() (in package ggpot2, Wickham 2016) has been used to produce the plot.
Survival seems to be worst for those patients with tumors with a histological
classification of adeno or smallcell.
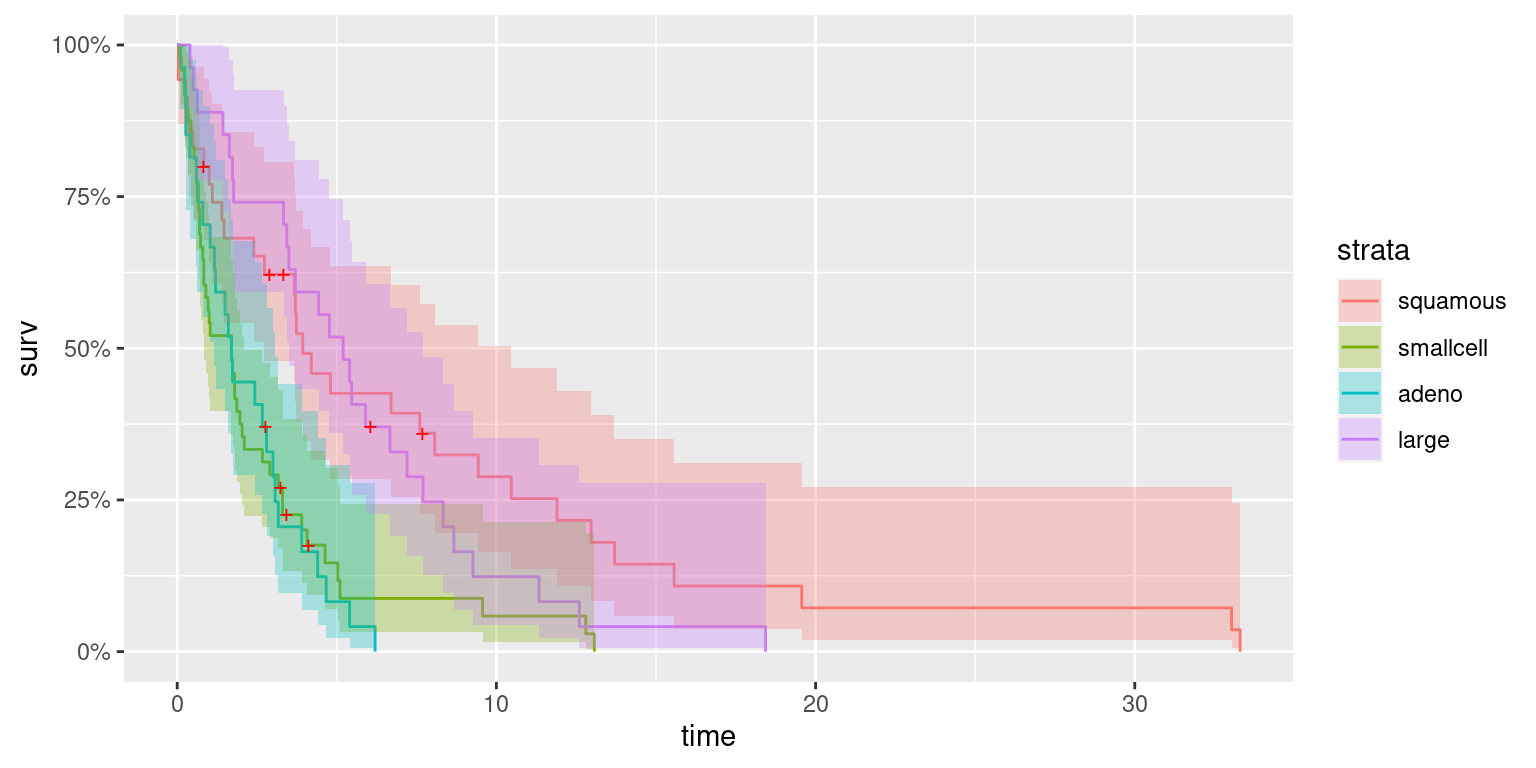
Figure 10.1: Survival estimates of patients according to histological type of tumor (i.e., cell type).
10.3 Parametric modeling of the survival function
INLA provides a number of distributions that can be used to model the
survival function, as seen in Table 10.2.
For this first example we will fit a survival model to the the veteran
dataset. INLA provides a similar function to Surv, called inla.surv, to
create the necessary data structure to conduct a survival analysis, which is
used as follows:
library("INLA")
sinla.vet <- inla.surv(veteran$time.m, veteran$status)The model fitted to the veteran dataset in this first example is an
exponential model. This corresponds to a model with constant hazard, i.e.,
\(h(t) = \lambda\), where \(\lambda\) is the rate parameter of the exponential
distribution (see Table 10.2).
exp.vet <- inla(sinla.vet ~ 1, data = veteran, family = "exponential.surv")
summary(exp.vet)##
## Call:
## "inla(formula = sinla.vet ~ 1, family = \"exponential.surv\", data
## = veteran)"
## Time used:
## Pre = 0.605, Running = 0.0532, Post = 0.0186, Total = 0.677
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -1.468 0.088 -1.646 -1.466 -1.298 -1.463 0
##
## Expected number of effective parameters(stdev): 1.00(0.00)
## Number of equivalent replicates : 136.89
##
## Marginal log-Likelihood: -317.37| Likelihood | \(h(t)\) | \(H(t)\) | \(S(t)\) | family |
|---|---|---|---|---|
| Exponential | \(\lambda\) | \(\lambda t\) | \(\exp(-\lambda t)\) | exponential.surv |
| Weibull | \(\alpha \lambda(\lambda t)^{\alpha -1}\) | \((\lambda t)^{\alpha}\) | \(\exp(-(\lambda t)^{\alpha})\) | weibull.surv |
| Loglogistic | \(\frac{(\beta/\alpha)(t/\alpha)^{\beta-1}}{1+(t/\alpha)^{\beta}}\) | - | \((1+(t/\alpha)^{\beta})^{-1}\) | loglogistic.surv |
| Lognormal | - | - | - | lognormal.surv |
In the previous model, a log link is used. Hence, \(\lambda = \exp(\alpha)\), where \(\alpha\) is the model intercept.
In this particular case, the survival function is
\[ S(t) = \exp(-H(t)) = \exp(-\int_0^t \lambda du) = \exp(-\lambda t) \]
We can use function inla.tmarginal() to transform the posterior marginal of
\(\alpha\) into the marginal of \(S(t)\) for a given time and compute 95% credible
intervals. Note that this is required because the intercept \(\alpha\) reported
by INLA is actually \(\log(\lambda)\). Hence, for each time \(t\), the marginal
of \(\alpha\) is transformed into the posterior marginal of \(S(t)\) and summary
statistics are computed with function inla.zmarginal():
library("parallel")
# Set number of cores to use
options(mc.cores = 4)
# Grid of times
times <- seq(0.01, 35, by = 1)
# Marginal of alpha
alpha.marg <- exp.vet$marginals.fixed[["(Intercept)"]]
# Compute post. marg. of survival function
S.inla <- mclapply(times, function(t){
S.marg <- inla.tmarginal(function(x) {exp(- exp(x) * t)}, alpha.marg)
S.stats <- inla.zmarginal(S.marg, silent = TRUE)
return(unlist(S.stats[c("quant0.025", "mean", "quant0.975")]))
})
S.inla <- as.data.frame(do.call(rbind, S.inla))Figure 10.2 shows the survival function obtained with an exponential model. Median survival time is about 4 months, similar to the value obtained with the non-parametric Kaplan-Meier estimator. Furthermore, uncertainty is very similar between both estimates, with the credible intervals of the Bayesian parametric estimate narrower in the sides.
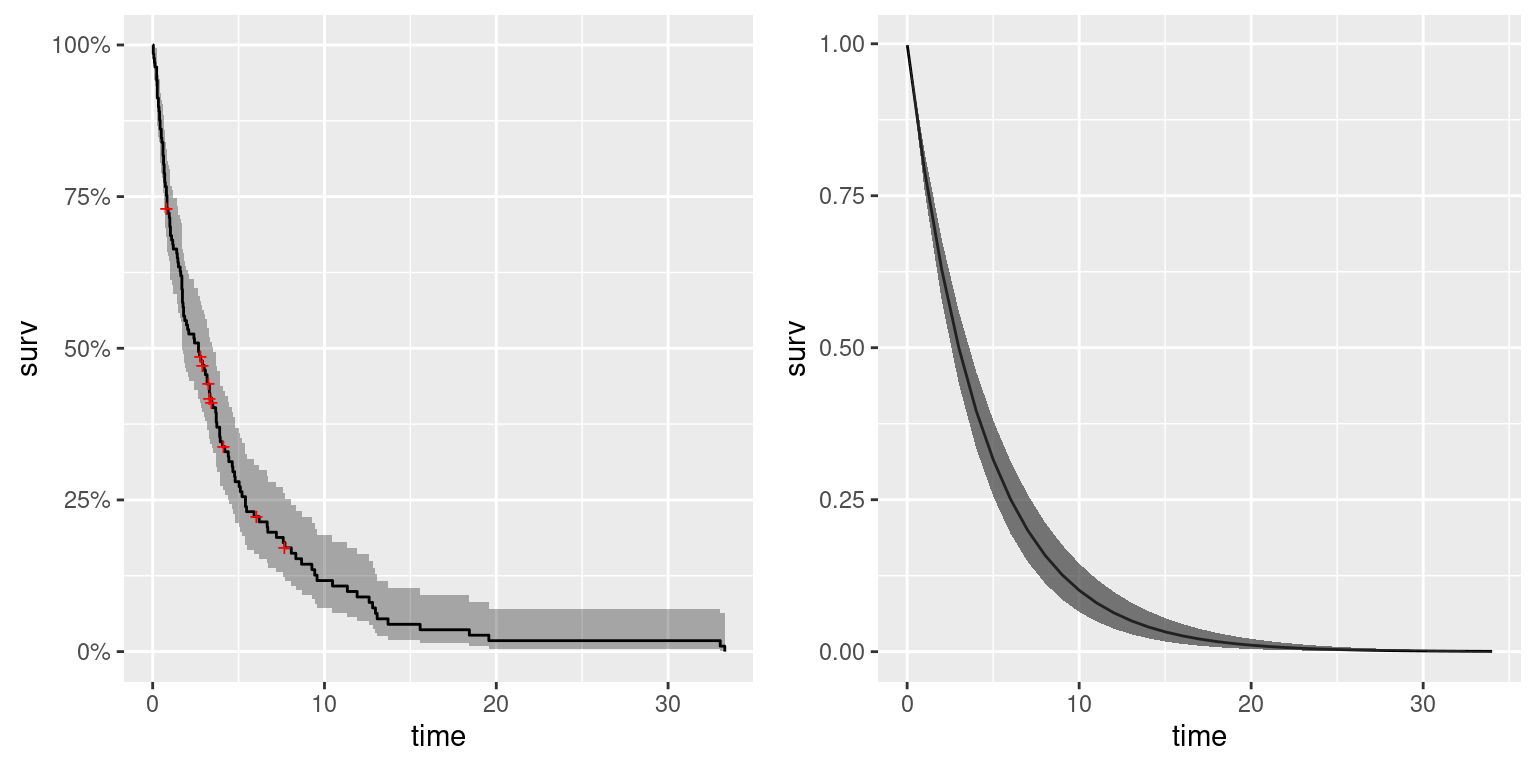
Figure 10.2: Kaplan-Meier estimate (left) and survival function using an exponential model (right) for the veteran dataset.
10.4 Semi-parametric estimation: Cox proportional hazards
Cox (1972) proposed a model in which the hazard function is the product of a baseline hazard \(h_0(t)\) and a term that depends on a number of covariates \(\mathbf{x}\). The baseline hazard can be estimated using non-parametric methods, while the term on the covariates is a function on a linear predictor on the covariates. In particular, a Cox proportional hazards models can be written down as:
\[ h(t) = h_0(t) \exp(\beta \mathbf{x}) \]
Hence, the hazard function is modulated by the covariates and their associated influence (as measured by coeffients \(\beta\)).
INLA implements a likelihood for a Cox proportional hazards model as follows:
\[ h(t) = h_0 (t) \exp(\eta) \]
Here, \(h_0(t)\) is the baseline hazard function and \(\eta\) is a linear predictor on some covariates and, possibly, other effects. Furthermore, the baseline hazard \(h_0(t)\) is modeled as:
\[ h_0(t) = \exp(b_k); t\in (s_{k-1}, s_{k}], k = 1, \ldots, K \]
Hence, the baseline hazard is a piecewise constant function. Variables \(\mathbf{b} = (b_1, \ldots, b_K)\) are assigned a prior which is a random walk of order 1 (default) or 2.
As described earlier, the veteran dataset includes a covariate that measures
whether the patient received a standard treatment or a test treatment. We will
use a Cox model to model survival time on this covariate and the type of
tumor in order to assess the effect of the test treatment.
Control option control.hazard (see Section 2.5) can
be used to define a number of options on how the baseline hazard function
is computed. Below, we only set the prior of the random effect to a
vague prior (as seen in Chapter 3).
coxinla.vet <- inla(sinla.vet ~ 1 + celltype + trt, data = veteran,
family = "coxph",
control.hazard = list(hyper = list(prec = list(param = c(0.001, 0.001)))))
summary(coxinla.vet)##
## Call:
## c("inla(formula = cph$formula, family = cph$family, contrasts =
## contrasts, ", " data = c(as.list(cph$data), cph$data.list),
## quantiles = quantiles, ", " E = cph$E, offset = offset, scale =
## scale, weights = weights, ", " Ntrials = NULL, strata = NULL,
## verbose = verbose, lincomb = lincomb, ", " selection = selection,
## control.compute = control.compute, ", " control.predictor =
## control.predictor, control.family = control.family, ", "
## control.inla = control.inla, control.results = control.results, ",
## " control.fixed = control.fixed, control.mode = control.mode, ", "
## control.expert = control.expert, control.hazard = control.hazard,
## ", " control.lincomb = control.lincomb, control.update =
## control.update, ", " control.pardiso = control.pardiso,
## only.hyperparam = only.hyperparam, ", " inla.call = inla.call,
## inla.arg = inla.arg, num.threads = num.threads, ", "
## blas.num.threads = blas.num.threads, keep = keep,
## working.directory = working.directory, ", " silent = silent, debug
## = debug)")
## Time used:
## Pre = 0.288, Running = 0.129, Post = 0.0229, Total = 0.44
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -2.194 0.263 -2.731 -2.187 -1.698 -2.174 0
## celltypesmallcell 1.147 0.260 0.641 1.145 1.663 1.141 0
## celltypeadeno 1.216 0.279 0.664 1.217 1.761 1.220 0
## celltypelarge 0.324 0.280 -0.228 0.325 0.870 0.327 0
## trttest 0.169 0.194 -0.213 0.169 0.550 0.169 0
##
## Random effects:
## Name Model
## baseline.hazard RW1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant
## Precision for baseline.hazard 437.30 545.22 17.03 237.95 1977.19
## mode
## Precision for baseline.hazard 37.60
##
## Expected number of effective parameters(stdev): 6.15(1.04)
## Number of equivalent replicates : 52.98
##
## Marginal log-Likelihood: -365.47The summary of the model indicates that survival depends on the type of tumor and that the test treatment does not seem to be better than the standard treatment.
The summary shows an estimate of the precision of the baseline hazard, which is modeled as a random walk of order 1 (in the log-scale). Because the estimate of the baseline risk is in the log-scale, we need to back-transform it and re-compute the summary statistics from the marginals:
#Transform baseline hazard
h0 <- lapply(coxinla.vet$marginals.random[["baseline.hazard"]],
function(X) {inla.tmarginal(exp, X)}
)
#Compute summary statistics for plotting
h0.stats <- lapply(h0, inla.zmarginal)Next, we have included the times and summary statistics of the baseline hazard together into a single data frame for plotting:
h0.df <- data.frame(t = coxinla.vet$summary.random[["baseline.hazard"]]$ID)
h0.df <- cbind(h0.df, do.call(rbind, lapply(h0.stats, unlist)))Finally, Figure 10.3 shows the posterior means and 95% credible intervals of the baseline hazard function. We have added a red line to mark a hazard of 1. The estimated baseline hazard function seems to be constant about 1. This may occur because the covariates included in the model explain most of the variability in the data.
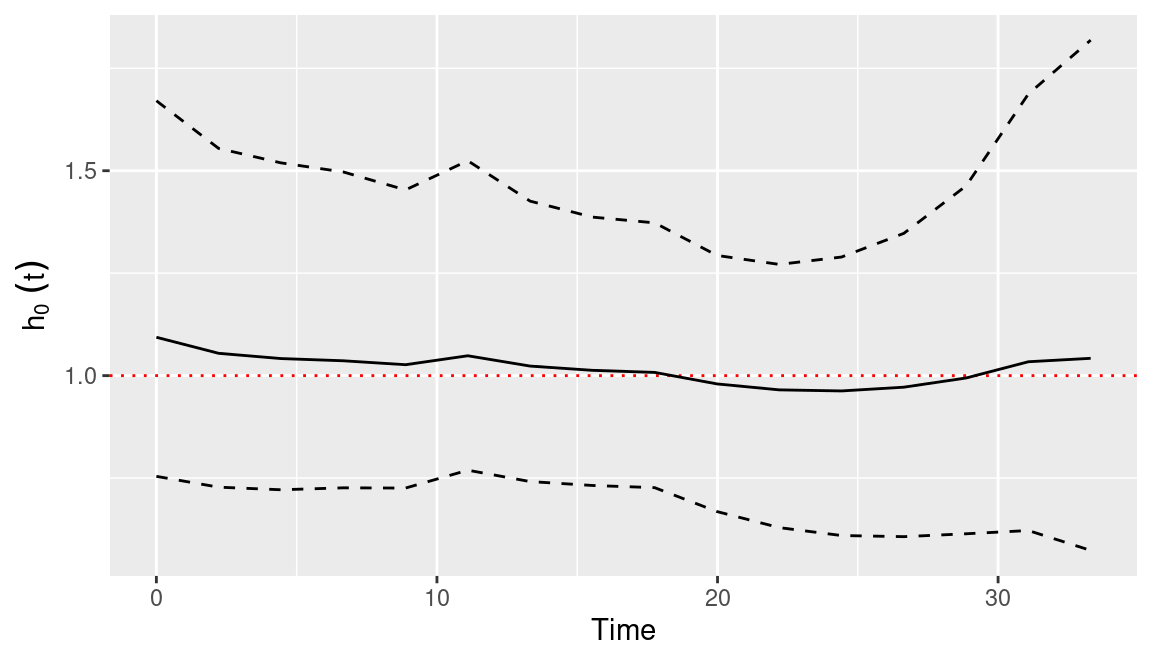
Figure 10.3: Posterior mean (solid line) and 95% credible interval (dashed lines) of baseline hazard function estimate using a Cox model on the veteran dataset.
10.5 Accelerated failure time models
Accelerated failure time (AFT) models are log-linear regression models that are widely applied in survival analysis. They take the following general form:
\[ \log(T) = \beta \mathbf{x} + \sigma \varepsilon \]
Here, \(\mathbf{x}\) is a vector of covariates with associated coefficients \(\beta\), \(\sigma\) is a precision parameter and \(\varepsilon\) is an error term. This error term can take different density distributions \(f_{\varepsilon}\) that will eventually define the survival and hazard functions.
| Distribution of \(T\) | Distribution of error term (\(f_{\varepsilon}\)) |
|---|---|
| Log-normal | Standard normal |
| Log-logistic | Logistic |
| Exponential | Extreme Value |
| Weibull | Extreme Value |
Table 10.3 shows the distributions for the error term \(\varepsilon\) and the corresponding distribution of \(T\).
10.5.1 Log-normal model
The way to specify the AFT model to use with INLA is via the family option.
In the next lines, a log-normal likelihood is used to fit a survival model
to the veteran dataset:
logninla.vet <- inla(sinla.vet ~ -1 + celltype + trt, data = veteran,
family = "lognormal.surv")Note that the log-survival likelihood used in the model (i.e.,
lognormal.surv) is different from the typical log-normal distribution (i.e.,,
lognormal), which does not takes censoring status into account.
The effects of the covariates on hazard can be assessed by checking the posterior summary statistics:
summary(logninla.vet)##
## Call:
## c("inla(formula = sinla.vet ~ -1 + celltype + trt, family =
## \"lognormal.surv\", ", " data = veteran)")
## Time used:
## Pre = 0.204, Running = 0.071, Post = 0.0185, Total = 0.293
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## celltypesquamous 1.254 0.266 0.734 1.254 1.779 1.252 0
## celltypesmallcell 0.420 0.211 0.005 0.419 0.835 0.419 0
## celltypeadeno 0.507 0.299 -0.080 0.506 1.094 0.506 0
## celltypelarge 1.495 0.276 0.953 1.495 2.038 1.494 0
## trttest -0.214 0.234 -0.674 -0.214 0.245 -0.214 0
##
## Model hyperparameters:
## mean sd 0.025quant
## Precision for the lognormalsurv observations 0.584 0.074 0.448
## 0.5quant 0.975quant mode
## Precision for the lognormalsurv observations 0.581 0.738 0.574
##
## Expected number of effective parameters(stdev): 5.00(0.00)
## Number of equivalent replicates : 27.40
##
## Marginal log-Likelihood: -342.10Given this model’s survival time, positive coefficients
indicate a longer survival. For example, the effect of having an
histological tumor of type large seems to provide the best survival
among all patients.
In order to assess what risk factors increase survival time (and reduce
hazard), the posterior probability of the coefficients of the fixed effects
being positive can be computed with function inla.pmarginal():
lapply(logninla.vet$marginals.fixed, function(X){1 - inla.pmarginal(0, X)})## $celltypesquamous
## [1] 1
##
## $celltypesmallcell
## [1] 0.9764
##
## $celltypeadeno
## [1] 0.955
##
## $celltypelarge
## [1] 1
##
## $trttest
## [1] 0.1779Note that logninla.vet$marginals.fixed is a list with all the posterior
marginals of the fixed effects. In order to compute the posterior probability
of being lower than one, one minus the probability of being lower than zero is
computed with 1 - inla.pmarginal(0, X) (where X represents a given posterior
marginal).
10.5.2 Weibull model
Next, a Weibull survival model will be fitted to the veteran dataset:
weibinla.vet <- inla(sinla.vet ~ -1 + celltype + trt,
data = veteran, family = "weibull.surv",
control.family = list(variant=0))The option control.family = list(variant=0) is used to use the following
parameterisation for the distribution of time \(t\):
\[ f(t) = \alpha t^{\alpha - 1} \lambda \exp\{-\lambda t^{\alpha}\} \]
Then, the linear predictor \(\eta\) is linked to \(\lambda\) as \(\log(\lambda) = \eta\).
summary(weibinla.vet)##
## Call:
## c("inla(formula = sinla.vet ~ -1 + celltype + trt, family =
## \"weibull.surv\", ", " data = veteran, control.family =
## list(variant = 0))")
## Time used:
## Pre = 0.202, Running = 0.101, Post = 0.0187, Total = 0.322
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## celltypesquamous -2.101 0.272 -2.648 -2.096 -1.581 -2.088 0
## celltypesmallcell -0.956 0.175 -1.309 -0.953 -0.623 -0.946 0
## celltypeadeno -0.889 0.242 -1.382 -0.883 -0.431 -0.870 0
## celltypelarge -1.777 0.236 -2.256 -1.771 -1.329 -1.761 0
## trttest 0.167 0.194 -0.214 0.167 0.547 0.167 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant
## alpha parameter for weibullsurv 0.98 0.056 0.867 0.981 1.09
## mode
## alpha parameter for weibullsurv 0.994
##
## Expected number of effective parameters(stdev): 5.00(0.00)
## Number of equivalent replicates : 27.40
##
## Marginal log-Likelihood: -322.59Because of the parameterisation of this model, negative coefficients will increase survival time (and reduce hazard). Hence, the posterior probability of the coefficients being lower than zero can be computed to assess increased hazard:
lapply(weibinla.vet$marginals.fixed, function(X){inla.pmarginal(0, X)})## $celltypesquamous
## [1] 1
##
## $celltypesmallcell
## [1] 1
##
## $celltypeadeno
## [1] 1
##
## $celltypelarge
## [1] 1
##
## $trttest
## [1] 0.195710.6 Frailty models
Under some circumstances, survival may be similar for certain patients. For example, members of a family may be resistant to certain types of cancer or pieces manufactured at a particular factory may have a lower quality that decreases survival time. This dependence means that observations are no longer independent within groups. This correlation is often modeled by means of random effects or frailties, in the survival argot.
Frailty models are essentially mixed-effects models for survival data.
Sometimes survival data are clustered or grouped (i.e., families, schools,
etc.) and observations are not independent. For this reason, random effects may
be included in the survival model to account for correlation between subjects
within groups.
In the next examples we will show how to use INLA to fit these types of
models.
The retinopathy dataset (Blair et al. 1976, @Husteretal:1989) in the survival
package records data from a trial on laser coagulation treatment to delay
diabetic retinopathy. This trial enrolled 197 patients with high-risk diabetic
retinopathy and the event of interest was the time of loss of vision (measured
as having a score below 5/200 in two consecutive visits). Each patient had one
eye treated and the other eye was used as a control. Variables in this dataset
are summarized in Table 10.4.
In this case, between patient variation will be modeled using random effects
in the frailty model.
| Variable | Description |
|---|---|
id |
Patient identification number. |
laser |
Type of laser used (‘xenon’ or ‘argon’). |
eye |
Eye that was treated (‘right’ or ‘left’). |
age |
Age at the diagnosis of diabetes. |
type |
Type of diabetes (‘juvenile’ or ‘adult’). |
trt |
Treatment (0 for control eye, 1 for treated eye). |
futime |
Time to loss of vision or last follow-up (in months). |
status |
Censoring status (0 for censored, 1 for loss of vision). |
risk |
Risk score for the eye (values of 6 or greater indicate high risk). |
First of all, the data will be loaded and the treatment variables converted into a factor:
data(retinopathy)
#Recode some covariates as factors
retinopathy$trt <- as.factor(c("control", "treated")[retinopathy$trt + 1])
#Summary
summary(retinopathy)## id laser eye age type
## Min. : 5 xenon:200 right:178 Min. : 1.0 juvenile:228
## 1st Qu.: 480 argon:194 left :216 1st Qu.:10.0 adult :166
## Median : 834 Median :16.0
## Mean : 873 Mean :20.8
## 3rd Qu.:1296 3rd Qu.:30.0
## Max. :1749 Max. :58.0
## trt futime status risk
## control:197 Min. : 0.3 Min. :0.000 Min. : 6.0
## treated:197 1st Qu.:14.0 1st Qu.:0.000 1st Qu.: 9.0
## Median :38.8 Median :0.000 Median :10.0
## Mean :35.6 Mean :0.393 Mean : 9.7
## 3rd Qu.:54.2 3rd Qu.:1.000 3rd Qu.:11.0
## Max. :75.0 Max. :1.000 Max. :12.0In order to avoid numerical problems, the time to event has been converted into years:
retinopathy$futime <- retinopathy$futime / 12A Kaplan-Meier estimate of the survival function can be obtained as:
surv.ret <- Surv(retinopathy$futime, retinopathy$status)
km.ret <- survfit(surv.ret ~ 1)Figure 10.4 shows the Kaplan-Meier estimate. The large amount of censored observations above 70 months can be seen, which makes it difficult to estimate median survival time and survival for large periods of time.
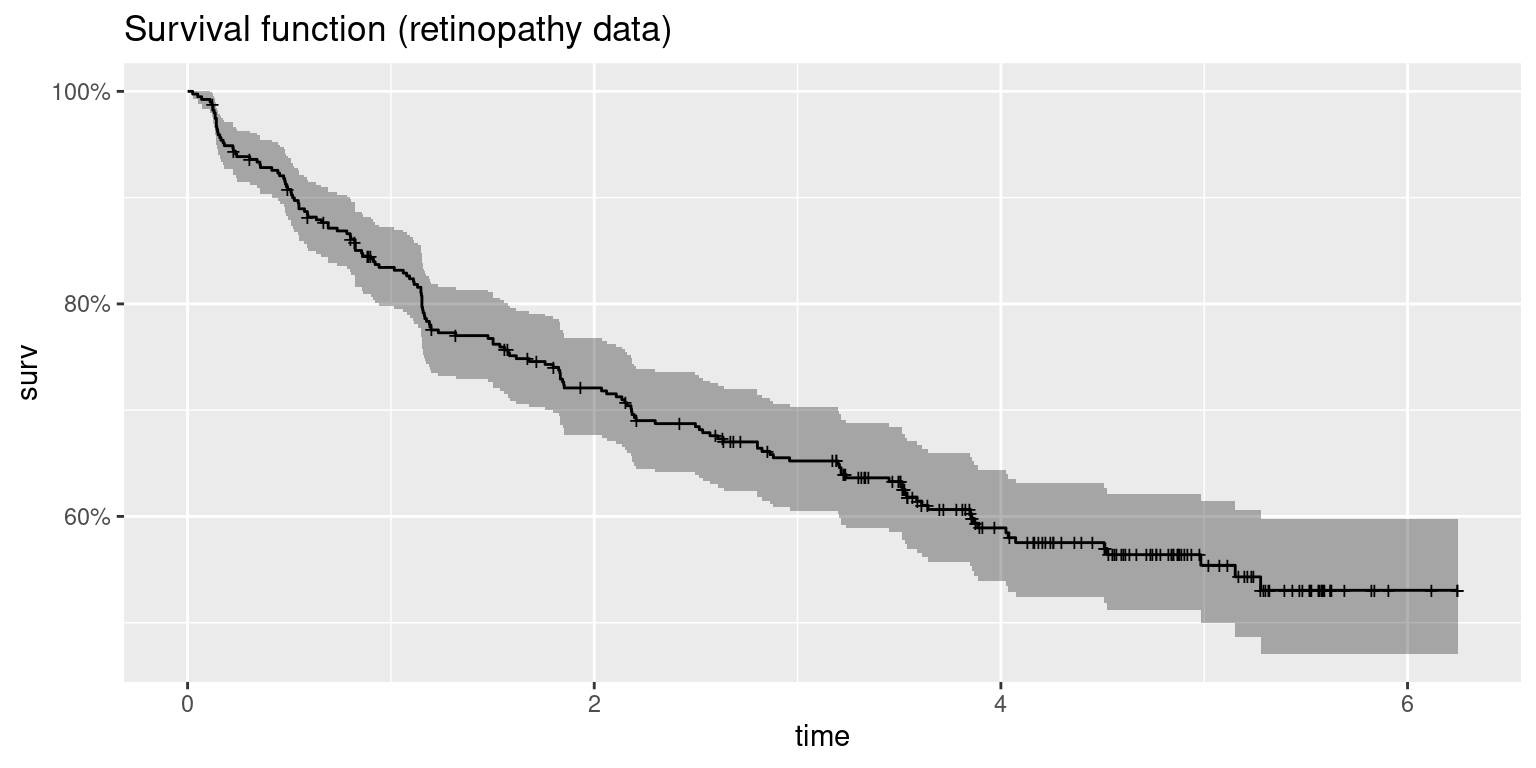
Figure 10.4: Survival function of the retinopathy dataset.
In this example we will consider a frailty model defined on patient (variable
id) and a vague prior is used on the precision of the frailty effect:
# Model formula
form <- inla.surv(futime, status) ~ 1 + trt + risk + f(id, model = "iid",
hyper = list(prec = list(param = c(0.001, 0.001))))
# Model fitting
fr.ret <- inla(form, data = retinopathy,
family = "weibullsurv")Note that in the previous model the random effects are defined using a Gaussian distribution. A summary of the model is:
summary(fr.ret)##
## Call:
## "inla(formula = form, family = \"weibullsurv\", data =
## retinopathy)"
## Time used:
## Pre = 0.227, Running = 0.45, Post = 0.0229, Total = 0.7
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -3.369 0.704 -4.783 -3.358 -2.019 -3.335 0
## trttreated -0.949 0.181 -1.310 -0.946 -0.599 -0.942 0
## risk 0.172 0.069 0.038 0.171 0.309 0.170 0
##
## Random effects:
## Name Model
## id IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant
## alpha parameter for weibullsurv 0.983 0.029 0.921 0.984 1.04
## Precision for id 1.204 0.491 0.585 1.090 2.46
## mode
## alpha parameter for weibullsurv 0.991
## Precision for id 0.905
##
## Expected number of effective parameters(stdev): 79.52(15.97)
## Number of equivalent replicates : 4.96
##
## Marginal log-Likelihood: -457.95The large precision of the frailties suggests that there is a small between patient variation given the other factors accounted for. Figure 10.5 shows 95% credible intervals of the frailties for each patient. These suggest that none of the patients have an intrinsic effect not accounted for by the covariates included in the model.
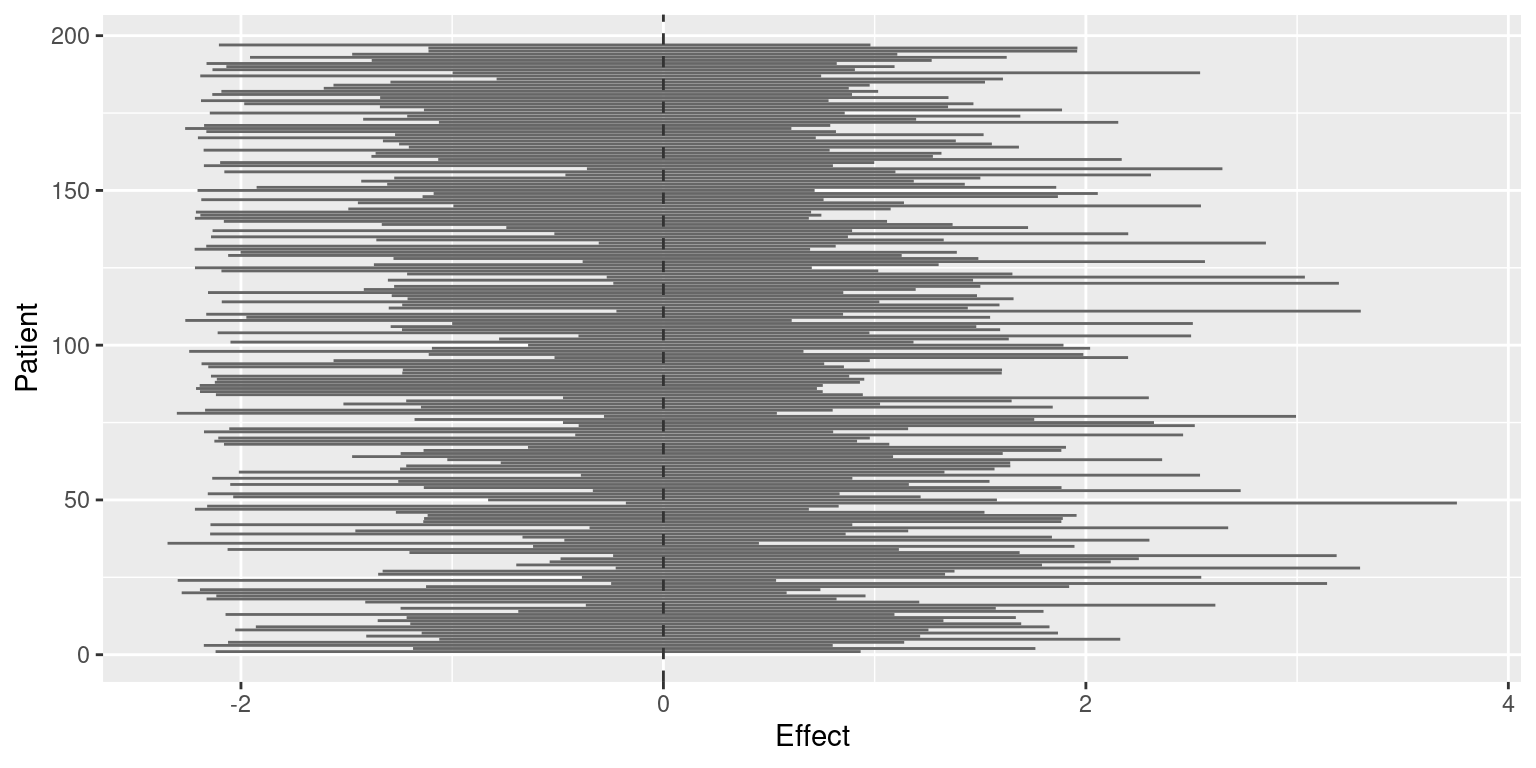
Figure 10.5: 95% credible intervals of the frailties in the retinopathy dataset analysis.
10.7 Joint modeling
So far, censorship and time to event have been assumed to be independent. However, these two variables may be correlated and relevant covariates may be available to explain this dependence. For example, patients may drop out from a clinical trial for a worsening in their clinical state. When clinical state can be modeled on several covariates using longitudinal data, censoring itself will also depend on these covariates. Hence, it is important to model clinical state (as measured by a convenient variable) and survival time.
This model will require a joint model with two components: a longitudinal model and a survival model. In practice, we will use a model with two likelihoods (see Section 6.4 for details) that share some latent effects. As an example, we will fit a joint model to a data set about 488 patients with liver cirrhosis in a randomized trial (Andersen et al. 1993). Prothrombin measurements were taken at irregular times as a measure of liver function. Furthermore, time to death (which can be censored) was also recorded for each patient. In addition, the randomized treatment for each patient (placebo or prednisone) is also available for all patients.
This dataset is available in package JMbayes (Rizopoulos 2016). The longitudinal and
survival parts of the data set is available as prothro, while the survival
part alone is prothros. In order to introduce the joint model step by step,
we will fit a longitudinal model first, followed by a purely survival model and
then the joint model.
10.7.1 Survival analysis
As stated before, the variables for the survival analysis are available in
dataset prothros in package JMbayes. Table 10.5 shows
a description of the variables in the dataset.
| Variable | Description |
|---|---|
id |
Patient id number. |
Time |
Time to event or censoring time. |
death |
Indicator of whether the patient died (1) or not (0). |
treat |
Treatment (placebo or prednisone). |
A summary of the variables in this dataset is shown below:
library("JMbayes")
data(prothros)
summary(prothros)## id Time death treat
## Min. : 1 Min. : 0.011 Min. :0.000 placebo :251
## 1st Qu.:167 1st Qu.: 0.632 1st Qu.:0.000 prednisone:237
## Median :302 Median : 2.616 Median :1.000
## Mean :295 Mean : 3.643 Mean :0.598
## 3rd Qu.:431 3rd Qu.: 6.236 3rd Qu.:1.000
## Max. :561 Max. :13.394 Max. :1.000A survival model of the patients with treatment as a covariate can be fit to
the data in order to assess the effect of the treatment and account for the
variability between the patients. Note that for survival models time to event
may be required to be re-scaled , e.g., to be in the \((0, 1)\) interval, to avoid
numerical problems with INLA. In this case, no re-scaling is done.
#prothros$Time <- prothros$Time / max(prothros$Time) # Re-scaling
pro.surv <- inla.surv(prothros$Time, prothros$death)
form.pro.surv <- pro.surv ~ 1 + treat
pro.surv.inla <- inla(form.pro.surv, data = prothros,
family = "weibull.surv")
summary(pro.surv.inla)##
## Call:
## "inla(formula = form.pro.surv, family = \"weibull.surv\", data =
## prothros)"
## Time used:
## Pre = 0.2, Running = 0.255, Post = 0.0167, Total = 0.472
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -1.561 0.106 -1.774 -1.560 -1.357 -1.557 0
## treatprednisone 0.093 0.117 -0.137 0.093 0.323 0.093 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant
## alpha parameter for weibullsurv 0.825 0.04 0.748 0.824 0.905
## mode
## alpha parameter for weibullsurv 0.823
##
## Expected number of effective parameters(stdev): 2.00(0.00)
## Number of equivalent replicates : 243.78
##
## Marginal log-Likelihood: -815.53The results show that prednisone has a positive coefficient but its effect does not seem to be significant, i.e., it does not increase survival time compared to the placebo.
10.7.2 Longitudinal analysis
Next, we will carry out a longitudinal analysis of the data to model how prothrombin measurements change over time. The longitudinal model will also consider the effect of treatment, which can be a placebo or prednisone.
| Variable | Description |
|---|---|
id |
Patient id number. |
pro |
Prothrombin measurement. |
time |
Time of prothrombin measurement. |
treat |
Treatment (placebo or prednisone). |
Time |
Time to event or censoring time. |
start |
Time of prothrombin measurement. |
stop |
Time of next prothrombin measurement or to event or censoring time. |
death |
Indicator of whether the patient died (1) or not (0). |
event |
Indicator of whether the event was observed in this time interval (1) or not (0). |
As stated earlier, the longitudinal data is available in dataset prothro in
package JMbayes. Table 10.6 describes the variables in the
longitudinal part of the dataset. Data is loaded and summarized below:
data(prothro)
summary(prothro)## id pro time treat
## Min. : 1 Min. : 6 Min. : 0.000 placebo :1498
## 1st Qu.:138 1st Qu.: 60 1st Qu.: 0.274 prednisone:1470
## Median :277 Median : 79 Median : 1.010
## Mean :277 Mean : 79 Mean : 2.039
## 3rd Qu.:418 3rd Qu.:100 3rd Qu.: 3.158
## Max. :561 Max. :176 Max. :11.108
## Time start stop death
## Min. : 0.011 Min. : 0.000 Min. : 0.011 Min. :0.00
## 1st Qu.: 2.467 1st Qu.: 0.274 1st Qu.: 0.539 1st Qu.:0.00
## Median : 5.632 Median : 1.010 Median : 1.851 Median :1.00
## Mean : 5.410 Mean : 2.039 Mean : 2.638 Mean :0.55
## 3rd Qu.: 8.145 3rd Qu.: 3.158 3rd Qu.: 4.082 3rd Qu.:1.00
## Max. :13.394 Max. :11.108 Max. :13.394 Max. :1.00
## event
## Min. :0.0000
## 1st Qu.:0.0000
## Median :0.0000
## Mean :0.0984
## 3rd Qu.:0.0000
## Max. :1.0000In this case, a longitudinal model is built to explain how prothrombin measurements change with time and according to treatment (as a fixed effect) or patient (included as a random effect). A log-normal likelihood is used in this case given that the response variable can only be positive:
form.pro.long <- pro ~ 1 + time + f(id, model = "iid") + treat
pro.long.inla <- inla(form.pro.long, data = prothro, family = "lognormal")
summary(pro.long.inla)##
## Call:
## "inla(formula = form.pro.long, family = \"lognormal\", data =
## prothro)"
## Time used:
## Pre = 0.24, Running = 1.81, Post = 0.0375, Total = 2.08
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 4.264 0.021 4.223 4.264 4.306 4.264 0
## time 0.018 0.003 0.013 0.018 0.023 0.018 0
## treatprednisone -0.093 0.030 -0.151 -0.093 -0.034 -0.093 0
##
## Random effects:
## Name Model
## id IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the lognormal observations 12.24 0.349 11.57 12.24
## Precision for id 11.39 0.918 9.69 11.35
## 0.975quant mode
## Precision for the lognormal observations 12.94 12.23
## Precision for id 13.30 11.27
##
## Expected number of effective parameters(stdev): 400.65(5.89)
## Number of equivalent replicates : 7.41
##
## Marginal log-Likelihood: -13745.93In this case, prothrombin increases with time but now patients treated with prednisone had lower values of prothrombin than patients treated with the placebo.
10.7.3 Joint model
For the joint model, we will be using a model with two likelihoods (see Section 6.4 for details), for the longitudinal and survival parts, respectively. First of all, we need to prepare the data. Using joint models with INLA is a bit tricky and requires extra preparation of the data.
First, we will create some auxiliary variables that we will use later.
Essentially, these variables will help us fill the empty spaces of the
variables used in the joint models with NA or zero.
#Number of observations in each dataset
n.l <- nrow(prothro)
n.s <- nrow(prothros)
#Vector of NA's
NAs.l <- rep(NA, n.l)
NAs.s <- rep(NA, n.s)
#Vector of zeros
zeros.l <- rep(0, n.l)
zeros.s <- rep(0, n.s)The longitudinal and survival responses, now called Y.long and Y.surv, need
to be expanded with NAs so that both variables have the same length
and then put together in a list:
#Long. and survival responses
Y.long <- c(prothro$pro, NAs.s)
Y.surv <- inla.surv(time = c(NAs.l, prothros$Time),
event = c(NAs.l, prothros$death))
Y.joint <- list(Y.long, Y.surv)Covariates need to be expanded in a similar way:
#Covariates
covariates <- data.frame(
#Intercepts (as factor with 2 levels)
inter = as.factor(c(rep("b.l", n.l), rep("b.s", n.s))),
#Time
time.l = c(prothro$time, NAs.s),
time.s = c(NAs.l, prothros$Time),
#Treatment (longitudinal)
treat.l = c(as.integer(prothro$treat =="prednisone"), NAs.s),
#Treat.s
treat.s = c(NAs.l, as.integer(prothros$treat =="prednisone"))
)
#Indices for random effects
r.effects <- list(
#Patiend id (long.)
id.l = c(prothro$id, NAs.s),
#Patient id (surv.)
id.s = c(NAs.l, prothros$id)
)The last step in the data preparation step is to create a new object,
joint.data, with the covariates, the indices of the random effects and
the response variables:
joint.data <- c(covariates, r.effects)
joint.data$Y <- Y.jointReferences
Andersen, P. K., O. Borgan, R. D. Gill, and N. Keiding. 1993. Statistical Models Based on Counting Processes. New York: Springer.
Blair, A. L., D. R. Hadden, J. A. Weaver, D. B. Archer, P. B. Johnston, and C. J. Maguire. 1976. “The 5-Year Prognosis for Vision in Diabetes.” American Journal of Ophthalmology 81: 383–96.
Cox, D. R. 1972. “Regression Models and Life-Tables.” Journal of the Royal Statistical Society. Series B 34 (2): 187–220.
Ibrahim, Joseph G., Ming-Hui Chen, and Debajyoti Sinha. 2001. Bayesian Survival Analysis. Springer.
Kalbfleisch, D., and R. L. Prentice. 1980. The Statistical Analysis of Failure Time Data. New York: Wiley.
Moore, Dirk F. 2016. Applied Survival Analysis Using R. Springer.
Rizopoulos, Dimitris. 2016. “The R Package JMbayes for Fitting Joint Models for Longitudinal and Time-to-Event Data Using Mcmc.” Journal of Statistical Software 72 (7): 1–45. https://doi.org/10.18637/jss.v072.i07.
Terry M. Therneau, and Patricia M. Grambsch. 2000. Modeling Survival Data: Extending the Cox Model. New York: Springer.
Therneau, Terry M. 2015. A Package for Survival Analysis in S. https://CRAN.R-project.org/package=survival.
Wickham, Hadley. 2016. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. https://ggplot2.tidyverse.org.