Chapter 3 Mixed-effects Models
3.1 Introduction
In Chapter 2 we have already introduced how to fit models with
fixed and random effects. In this chapter a more detailed description of the
different types of fixed and random effects available in INLA will be
provided.
First of all, let’s recall that a covariate should enter the model as a linear fixed effect when it is thought that it affects all observations in the same way (in a linear way, in fact) and that this effect is of primary interest in the study.
Random effects, on the other hand, are often adopted to take into account variation produced by variables in the model that are not our primary object of study. For example, in a study to investigate the effect of different irrigation systems on plant growth, measurement may include the type of irrigation, type of soil and variables associated with the trees, such as a numerical identifier of the tree, age, etc. Trees themselves in the study are not the primary object of study as they are a (random) sample of the population of trees. However, depending on the actual tree being measured, growth may be faster or slower and it will affect the growth measurement. In this case, because the trees in the study are taken as a random sample from the total population of trees, their effect is included as a random effect.
In a Bayesian context, a fixed effect will have an associated coefficient which is often assigned a vague prior, such as a Gaussian with zero mean and large variance. On the other hand, random effects will have a common Gaussian prior with a zero mean and a precision, to which a prior will be assigned.
3.2 Fixed-effects models
As explained in Section 2.3, fixed effects can be easily included
in the model formula. The default prior assigned to the associated coefficients
(and the intercept) is a Gaussian distribution, and its parameters can be set
through option control.fixed in the call to inla().
Fixed effects can also be included in the model by including one or more terms
in the model formula using the the f() function. Table
3.1 summarizes the two types of fixed effects implemented
in INLA, which will be described in more detail below.
| Name | Description |
|---|---|
linear |
Linear fixed effect. |
clinear |
Linear fixed effect with constrained coefficient. |
3.2.1 linear fixed effect
The linear effect is similar to including a covariate in the formula in
the usual way and it implements the effect \(\beta x_i\), where \(x_i\) is
the value of the covariate for observation \(i\) and \(\beta\) the associated
coefficient. The prior on \(\beta\) is a Gaussian distribution with
mean and precision that can be specified in the call to the f() function
when the model is defined in the formula. Table 3.2
summarizes the specific options available for this effect. Alternatively,
parameter control.fixed can be used to set the prior on the
coefficient.
| Argument | Default | Description |
|---|---|---|
mean.linear |
\(0\) | Mean of Gaussian prior on a coefficient. |
prec.linear |
\(0.001\) | Precision of Gaussian prior on a coefficient. |
3.2.2 clinear fixed effect
Sometimes it is necessary to fit a model in which the covariate coefficients
are constrained in some way, e.g., we want to force the coefficient to be
non-negative. This can be achieved by means of the clinear
effect. Table 3.3 shows the different options
available to specify this effect. Note that precision is the precision
of a (tiny) error that is added to the actual value of \(\beta x_i\) required
by how this latent effect is implemented.
| Argument | Default | Description |
|---|---|---|
range |
c(-Inf, Inf) |
Vector of length two with the range of the linear coefficient. |
precision |
exp(14) |
Tiny precision to implement this latent effect. |
The implementation of this effect distinguishes three different
possibilities depending on how range is defined:
range = c(-Inf, Inf)In this case there are not constraints and the model is equivalent to the
lineareffect. The parameterization is \(\beta = \theta\), where \(\theta\) is the parameter in the internal scale used byINLAto estimate the model.range = c(a, Inf)In this case
ais the lower bound of the parameter and there is no upper limit, so the internal parameterization is\[ \beta = a + \exp(\theta) \]
range = c(a, b)This is the model general case in which both limits, lower bound
aand upper boundb, are real numbers. The internal parameterization is now:\[ \beta = a + (b - a) \frac{\exp(\theta)}{1 + \exp(\theta)} \]
In all cases the prior of this model is set on the internal parameter \(\theta\) and it is a Gaussian distribution with default mean equal to 1 and precision equal to 10.
In order to illustrate the use of the two models for fixed effects we will revisit the example on the SIDS dataset. A model will be fitted to constrain to a positive coefficient on the covariate that represents the proportion of non-white births in a county.
First of all, the dataset will be loaded and the required variables computed (see Chapter 2 for details):
library("spdep")
data(nc.sids)
# Overall rate
r <- sum(nc.sids$SID74) / sum(nc.sids$BIR74)
nc.sids$EXP74 <- r * nc.sids$BIR74
# Proportion of non-white births
nc.sids$NWPROP74 <- nc.sids$NWBIR74 / nc.sids$BIR74Next, the two models will be fitted. First of all, the model
using the linear effect:
m.pois.lin1 <- inla(SID74 ~ f(NWPROP74, model = "linear"),
data = nc.sids, family = "poisson", E = EXP74)
summary(m.pois.lin1)##
## Call:
## c("inla(formula = SID74 ~ f(NWPROP74, model = \"linear\"), family
## = \"poisson\", ", " data = nc.sids, E = EXP74)")
## Time used:
## Pre = 0.352, Running = 0.0592, Post = 0.0201, Total = 0.432
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.646 0.090 -0.824 -0.645 -0.470 -0.644 0
## NWPROP74 1.869 0.217 1.440 1.869 2.293 1.870 0
##
## Expected number of effective parameters(stdev): 2.00(0.00)
## Number of equivalent replicates : 49.90
##
## Marginal log-Likelihood: -226.12Note how the posterior marginal of the coefficient of covariate NWPROP74 has
a 95% credible interval which is above zero. In case we wanted this coefficient
to be positive, this is how the model with a constraint on the coefficient
using the clinear effect is fitted:
library("INLA")
m.pois.clin1 <- inla(SID74 ~ f(NWPROP74, model = "clinear",
range = c(0, Inf)), data = nc.sids, family = "poisson",
E = EXP74)
summary(m.pois.clin1)##
## Call:
## c("inla(formula = SID74 ~ f(NWPROP74, model = \"clinear\", range =
## c(0, ", " Inf)), family = \"poisson\", data = nc.sids, E = EXP74)"
## )
## Time used:
## Pre = 0.211, Running = 0.0923, Post = 0.0211, Total = 0.324
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.672 0.084 -0.843 -0.67 -0.514 -0.666 0.002
##
## Random effects:
## Name Model
## NWPROP74 Constrained linear
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Beta for NWPROP74 1.93 0.20 1.54 1.93 2.33 1.93
##
## Expected number of effective parameters(stdev): 1.00(0.00)
## Number of equivalent replicates : 99.55
##
## Marginal log-Likelihood: -222.89It can be seen how the estimates of the intercept and the coefficient of the proportion of non-white births are very similar between the unconstrained and the constrained model.
3.3 Types of mixed-effects models
The list of random effects implemented in INLA is quite rich.
Random effects in INLA are defined using a multivariate Gaussian distribution
with zero mean and precision matrix \(\tau \Sigma\), where \(\tau\) is a generic
precision parameter and \(\Sigma\) is a matrix that defines the dependence
structure of the random effects and that may depend on further parameters.
When the random effects are a latent Gaussian Markov random field the structure
of \(\Sigma\) is quite sparse and this can be exploited for computational purposes
(Rue and Held 2005). Given a vector \(\mathbf{u}\) of random effects, this will be
represented as:
\[ \mathbf{u} \sim N(0, \tau \Sigma) . \]
Table 3.4 summarizes the ones described in this chapter, but other types of random effects are discussed in different chapters.
| Name | Description |
|---|---|
iid |
Independent and identically distributed Gaussian random effect. |
z |
“Classical” specification of random effects. |
generic0 |
Generic specification (type 0). |
generic1 |
Generic specification (type 1). |
generic2 |
Generic specification (type 2). |
generic3 |
Generic specification (type 3). |
rw1 |
Random walk of order 1. |
rw2 |
Random walk of order 2. |
crw2 |
Continuous random walk of order 2. |
seasonal |
Seasonal variation with a given periodicity. |
ar1 |
Autoregressive model of order 1. |
ar |
Autoregressive model of arbitrary order. |
iid1d |
Correlated random effects of dimension 1. |
iid2d |
Correlated random effects of dimension 2. |
iid3d |
Correlated random effects of dimension 3. |
iid4d |
Correlated random effects of dimension 4. |
iid5d |
Correlated random effects of dimension 5. |
3.3.1 Independent random effects (iid model)
Independent and identically distributed random effects have already been introduced in Chapter 2 and they are probably the simplest way to account for unstructured variability in the data.
The precision matrix of iid random effects is \(\tau \Sigma\) with \(\Sigma\) a
diagonal matrix of scaling factors \(\{s_1, \ldots,s_n\}\) (which are all equal
to 1 by default). Scaling factors are defined by means of parameter scale
within the f() function used to define the latent effect in the formula.
The internal parameterization of the hyperparameter of this model is \(\theta = \log (\tau)\), which by default is assigned a log-Gamma distribution with parameters \(1\) and \(0.00005\). In other words, the prior of \(\tau\) is a Gamma distribution.
The first argument passed to the f() function should be an index of values
that define the grouping of the observations in order to define the number of
random effects. The length of parameter scale, if used, must be equal to the
number of groups defined. Scaling may be used, for example, to account for
different sample sizes in the groups defined by the data (see Section
5.6).
This type of random effects is often used to account for overdispersion in Poisson models. For this reason, the example will be again based on the SIDS dataset. The first step is to create an index for the random effects:
nc.sids$ID <- 1:100Here, the index goes from 1 to 100 because we want a different value of the random effect for each county. If different counties must share the same random effect then the index variable should assign the same value to these counties.
The iid is then added to the model formula, using the f() function:
m.pois.iid <- inla(SID74 ~ f(ID, model = "iid"),
data = nc.sids, family = "poisson",
E = EXP74)
summary(m.pois.iid)##
## Call:
## c("inla(formula = SID74 ~ f(ID, model = \"iid\"), family =
## \"poisson\", ", " data = nc.sids, E = EXP74)")
## Time used:
## Pre = 0.211, Running = 0.143, Post = 0.0191, Total = 0.374
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.028 0.063 -0.155 -0.026 0.092 -0.024 0
##
## Random effects:
## Name Model
## ID IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Precision for ID 7.26 2.57 3.79 6.77 13.61 6.02
##
## Expected number of effective parameters(stdev): 40.44(6.52)
## Number of equivalent replicates : 2.47
##
## Marginal log-Likelihood: -245.54The posterior mean of the precision of the random effects is 7.2609, which is a clear sign of overdispersion.
The same model can now include the covariate on the proportion of non-white births:
m.pois.iid2 <- inla(SID74 ~ NWPROP74 + f(ID, model = "iid"),
data = nc.sids, family = "poisson",
E = EXP74)
summary(m.pois.iid2)##
## Call:
## c("inla(formula = SID74 ~ NWPROP74 + f(ID, model = \"iid\"),
## family = \"poisson\", ", " data = nc.sids, E = EXP74)")
## Time used:
## Pre = 0.212, Running = 0.117, Post = 0.0193, Total = 0.348
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.646 0.093 -0.829 -0.645 -0.466 -0.644 0
## NWPROP74 1.871 0.224 1.431 1.871 2.310 1.872 0
##
## Random effects:
## Name Model
## ID IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Precision for ID 15986.25 19485.14 14.00 9361.65 69186.70 15.69
##
## Expected number of effective parameters(stdev): 5.04(7.11)
## Number of equivalent replicates : 19.83
##
## Marginal log-Likelihood: -227.85Now, the posterior mean of the precision of the random effects is \(1.5986\times 10^{4}\). This means that the covariate explains most of the overdispersion in the data.
3.3.2 Generic specification
In general, iid random effects are not suitable to model complex covariate
structures that often appear in experimental design. For this reason, INLA
provides a number of generic specifications for random effects, in addition to
some specific ones (that are described later in this chapter).
3.3.3 Mixed-effects design matrix (z model)
The z latent model can be used to define a term \(Z \mathbf{u}\) in the linear
predictor where \(Z\) is a defined matrix and \(\mathbf{u}\) is a vector of iid
random effects with zero mean and precision \(\tau C\). \(C\) is a known matrix,
and it is taken to be a diagonal matrix of the appropriate dimension if it is
not specified.
This random effect allows for the following representation of the linear predictor:
\[ \eta = X \beta + Z \mathbf{u} \] which is a standard and generic representation of mixed-effects models (Pinheiro and Bates 2000). Hence, \(Z\) can be regarded as a “design matrix” for the random effects and it will allow one observation to depend on more than one random effect. Note that \(Z\) will have as many rows as observations and as many columns as elements in the vector of random effects \(\mathbf{u}\).
Because of the way this latent effect is implemented in INLA, the actual
model is:
\[ \eta = Z\mathbf{u} + \varepsilon \]
Here, \(\eta\) is the contribution of the z random effect to the linear
predictor and \(\varepsilon\) a tiny random error added. The precision of this
tiny error can be set when the random effect is defined. All the specific
options of this random effect that can be passed to the f() function are
shown in Table 3.5.
| Argument | Default | Description |
|---|---|---|
Z |
— | Mixed-effects design matrix \(Z\). |
Cmatrix |
Diagonal matrix | Structure of precision of random effects (\(C\) matrix). |
precision |
exp(14) |
Precision of tiny noise \(\varepsilon\). |
The next example shows how to fit the model with random effects on the SIDS
dataset using the z random effect:
m.pois.z <- inla(SID74 ~ f(ID, model = "z", Z = Diagonal(nrow(nc.sids), 1)),
data = nc.sids, family = "poisson", E = EXP74)
summary(m.pois.z)##
## Call:
## c("inla(formula = SID74 ~ f(ID, model = \"z\", Z =
## Diagonal(nrow(nc.sids), ", " 1)), family = \"poisson\", data =
## nc.sids, E = EXP74)")
## Time used:
## Pre = 0.237, Running = 0.264, Post = 0.0197, Total = 0.521
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.028 0.063 -0.155 -0.026 0.092 -0.024 0
##
## Random effects:
## Name Model
## ID Z model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Precision for ID 7.26 2.57 3.79 6.77 13.62 6.02
##
## Expected number of effective parameters(stdev): 40.44(6.52)
## Number of equivalent replicates : 2.47
##
## Marginal log-Likelihood: -245.54Note how the estimates of the intercept and the precision of the random
effects are very similar to the model fit before using the iid latent
effects.
3.3.4 Type 0 generic specification (generic0 model)
In this latent effect the value of matrix \(\Sigma\) is a fixed matrix \(C\) that
is completely known and does not depend on any other hyperparameter.
This is specified via the Cmatrix argument in the f() function
and it must be a non-singular symmetric matrix, so that a proper
precision matrix is defined.
This latent effect has a single hyperparameter precision \(\tau\), and the prior is set on \(\theta = \log(\tau)\) and it is, by default, a log-Gamma with parameters \(1\) (shape) and \(0.00005\) (rate).
We go back to the model with random effects using the SIDS dataset. In this case, we set matrix \(C\) to be diagonal as this defines the same model used in previous examples:
m.pois.g0 <- inla(SID74 ~ f(ID, model = "generic0",
Cmatrix = Diagonal(nrow(nc.sids), 1)),
data = nc.sids, family = "poisson", E = EXP74)
summary(m.pois.g0)##
## Call:
## c("inla(formula = SID74 ~ f(ID, model = \"generic0\", Cmatrix =
## Diagonal(nrow(nc.sids), ", " 1)), family = \"poisson\", data =
## nc.sids, E = EXP74)")
## Time used:
## Pre = 0.205, Running = 0.141, Post = 0.0191, Total = 0.365
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.028 0.063 -0.155 -0.026 0.092 -0.024 0
##
## Random effects:
## Name Model
## ID Generic0 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Precision for ID 7.26 2.57 3.79 6.77 13.61 6.02
##
## Expected number of effective parameters(stdev): 40.44(6.52)
## Number of equivalent replicates : 2.47
##
## Marginal log-Likelihood: -245.54Given than the fit model is the same one as in previous examples, the estimates of the intercept and the precision are very similar to those obtained above.
3.3.5 Type 1 generic specification (generic1 model)
This latent effect has a structure of the precision matrix which is \((I - \frac{\beta}{\lambda_{max}}C)\), where \(I\) is the diagonal matrix, \(C\) a known
matrix, \(\lambda_{max}\) its maximum eigenvalue and \(\beta\) a parameter in
the \([0,1)\) interval. Matrix \(C\) is passed to the f() function
using the Cmatrix argument.
The internal parameterization of this latent effect is \(\theta_1 = \log(\tau)\) and \(\theta_2 = \textrm{logit}(\beta)\). For \(\theta_1\) the default prior is a log-Gamma with parameters \(1\) and \(0.00005\), while for \(\theta_2\) is a Gaussian with zero mean and precision \(0.01\).
The generic1 latent effect has been used to implement some spatial models
(Bivand, Gómez-Rubio, and HRue 2014; Bivand, Gómez-Rubio, and Rue 2015) not included in the INLA package. An
example on the use of the generic1 latent effect is provided in Section
7.2 to fit the spatial model proposed in Leroux, Lei, and Breslow (1999).
3.3.6 Type 2 generic specification (generic2 model)
This latent effect is used to represent the following hierarchical model:
\[ v \sim N(0, \tau_v C) \]
\[ u \mid v \sim N(v, \tau_u I) \]
and results in the following structure for the precision matrix:
\[ Q = \left[ \begin{array}{cc} \tau_u I & -\tau_u I\\ -\tau_u I & \tau_u I + \tau_v C\\ \end{array} \right] \]
Hence, the resulting vector of random effects is \((u, v)\). Matrix \(C\) is
passed to the f() function using the Cmatrix argument.
The priors are set on internal parameters \(\theta_1 = \log(\tau_v)\) and \(\theta_2 = \log(\tau_u)\) By default, they are both a log-Gamma with parameters \(1\) and \(0.00005\), and \(1\) and \(0.001\), respectively.
3.3.7 Type 3 generic specification (generic3 model)
The last form of generic random effect has a precision matrix that is a linear combination of \(K\)different terms:
\[ Q = \tau \sum_{i=1}^K \tau_i C_i \]
Here, \(\tau\) is a common precision parameter, \(\tau_i\) a term-specific
precision parameter associated with matrix \(C_i\). \(K\) is the number of
terms in the sum, and it must be an integer value from 1 to 10. Matrices \(C_i\)
are passed using a list with argument Cmatrix in the definition of the latent
effect in the model formula.
Priors are set on parameters \(\theta_1, \ldots, \theta_{11}\), with \(\theta_i = \log(\tau_i), i = 1,\ldots, 10\) and \(\theta_{11} = \log(\tau)\). By default, priors on \(\theta_i,\ i=1,\ldots,10\) are log-Gamma with parameters \(1\) and \(0.00005\). Parameter \(\theta_{11}\) is fixed to zero by default (i.e., \(\tau = 1\) by default).
Note that in order to make this model identifiable only \(K\) precision
parameters can be free out of the \(K+1\) in the model. For this reason, \(\tau\)
is fixed by default to a fixed value of 1 (0 in the log-scale). This parameter
could be set to a different fixed value to rescale all other precision
parameters, which sometimes help to identify the model parameters better. See
also Section 5.6 for a general discussion on scaling of
hyperparameters in INLA.
In the following example we consider the SIDS dataset again and fit a Poisson
model with a random effect for overdispersion using the generic3 latent
effect. First, a diagonal matrix K1 is defined and then a list
with this matrix is used to define the latent effect:
K1 <- Diagonal(nrow(nc.sids), 1)
m.pois.g2 <- inla(SID74 ~ f(ID, model = "generic3", Cmatrix = list(K1)),
data = nc.sids, family = "poisson", E = EXP74)
summary(m.pois.g2)##
## Call:
## c("inla(formula = SID74 ~ f(ID, model = \"generic3\", Cmatrix =
## list(K1)), ", " family = \"poisson\", data = nc.sids, E = EXP74)")
## Time used:
## Pre = 0.227, Running = 0.148, Post = 0.0192, Total = 0.394
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.028 0.063 -0.155 -0.026 0.092 -0.024 0
##
## Random effects:
## Name Model
## ID Generic3 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant
## Precision for Cmatrix[[1]] for ID 7.26 2.57 3.79 6.77 13.61
## mode
## Precision for Cmatrix[[1]] for ID 6.02
##
## Expected number of effective parameters(stdev): 40.44(6.52)
## Number of equivalent replicates : 2.47
##
## Marginal log-Likelihood: -245.54As expected, the estimates of the intercept and precision are equal to the ones obtained when the same model was fitted using other random effects.
3.3.9 Random walks (rw1 and rw2 models)
Random walks of order one and two are also available as latent
effects in INLA. Given a vector of Gaussian observations
\(u = (u_1,\ldots, u_n)\), the random walk of order one is defined
assuming that increments \(\Delta u_i = u_{i} - u_{i-1}\) follow
a Gaussian distribution with zero mean and precision \(\tau\).
This is equivalent to assuming that the distribution of
vector \(u\) is Gaussian with zero mean and precision matrix
\(\tau Q\), where \(Q\) contains the neighborhood structure of the
model.
Similarly, the random walk of order two is defined by assuming that second order increments \(\Delta^2 u_i = u_{i} - 2u_{i-1} + u_{i-2}\) follow a Gaussian distribution with zero mean and precision \(\tau\).
In both cases the random walk can be defined to be cyclic, and the model can be
scaled to have an average variance (i.e., the diagonal of the generalized
inverse of \(Q\)) equal to 1 (see Section 5.6). See Table
3.7 for options cyclic and scale.model, respectively.
Although this is a discrete latent effect, it can be used to model continuous
variables by using the values argument. In particular, the values of the
covariate can be binned so that they are associated with a particular value
of the vector of random effects. This is particularly interesting to model
non-linear dependence on covariates in the linear predictor.
A continuous random walk of order 2 is implemented as model crw2 in INLA.
See Chapter 3 in Rue and Held (2005) for details on how this model is defined as a
latent Gaussian Markov random field. In particular, these models are consistent
with respect to the choice of the locations and the resolution, and their
precision matrix is sparse due to the fact that they fulfill a Markov property.
| Argument | Default | Description |
|---|---|---|
values |
— | Values of the covariate for which the effect is estimated. |
cyclic |
FALSE |
Cyclic adjacency. |
scale.model |
FALSE |
Whether to scale variance matrix. |
The AirPassengers dataset records the number of total monthly international
airline passengers from 1949 to 1960 (Box, Jenkins, and Reinsel 1976). This data is an object of
class ts (i.e., time-series) and the next lines of R code show how to load
the data and create a data.frame with the number of passengers, year, month
of the year and id to identify the row in the dataset:
data(AirPassengers)
airp.data <- data.frame(airp = as.vector(AirPassengers),
month = as.factor(rep(1:12, 12)),
year = as.factor(rep(1949:1960, each = 12)),
ID = 1:length(AirPassengers))Figure 3.2 shows the original times series and the log of the number of passengers. The original data show an increasing variance with time, which is not observed in the log-transformed time series. Given that the random walk assumes a common precision, it will be better to model the data in the log-scale.
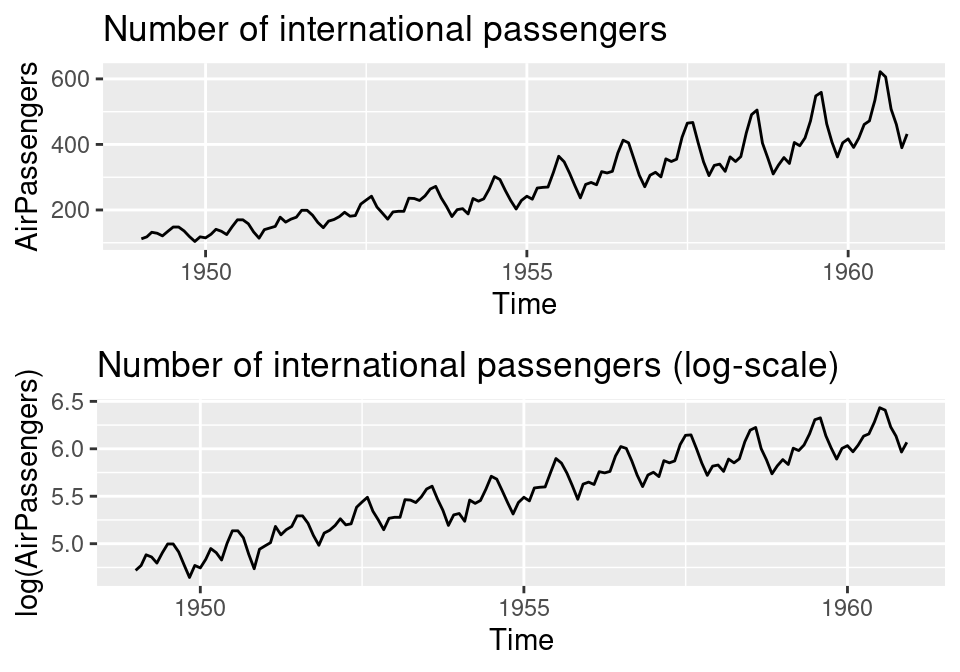
Figure 3.2: Monthly international airline passengers: original data (top) and log-scale (bottom).
The first model to be fit to the data is a linear model with a
fixed effect on year and a rw1 random effect:
airp.rw1 <- inla(log(AirPassengers) ~ 0 + year + f(ID, model = "rw1"),
control.family = list(hyper = list(prec = list(param = c(1, 0.2)))),
data = airp.data, control.predictor = list(compute = TRUE))
summary(airp.rw1)##
## Call:
## c("inla(formula = log(AirPassengers) ~ 0 + year + f(ID, model =
## \"rw1\"), ", " data = airp.data, control.predictor = list(compute
## = TRUE), ", " control.family = list(hyper = list(prec = list(param
## = c(1, ", " 0.2)))))")
## Time used:
## Pre = 0.328, Running = 0.573, Post = 0.112, Total = 1.01
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## year1949 5.157 0.252 4.664 5.156 5.654 5.154 0
## year1950 5.207 0.220 4.776 5.206 5.641 5.205 0
## year1951 5.332 0.190 4.959 5.332 5.707 5.331 0
## year1952 5.420 0.165 5.096 5.420 5.745 5.420 0
## year1953 5.490 0.146 5.203 5.490 5.776 5.490 0
## year1954 5.517 0.135 5.251 5.517 5.782 5.517 0
## year1955 5.602 0.135 5.335 5.602 5.867 5.603 0
## year1956 5.670 0.146 5.381 5.670 5.956 5.671 0
## year1957 5.727 0.165 5.401 5.728 6.051 5.729 0
## year1958 5.736 0.190 5.360 5.736 6.109 5.737 0
## year1959 5.806 0.219 5.374 5.807 6.237 5.807 0
## year1960 5.843 0.251 5.348 5.843 6.336 5.844 0
##
## Random effects:
## Name Model
## ID RW1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 100.92 18.11 69.24 99.68
## Precision for ID 157.70 42.63 93.83 150.91
## 0.975quant mode
## Precision for the Gaussian observations 139.98 97.46
## Precision for ID 259.68 138.00
##
## Expected number of effective parameters(stdev): 58.90(8.78)
## Number of equivalent replicates : 2.44
##
## Marginal log-Likelihood: 2.04
## Posterior marginals for the linear predictor and
## the fitted values are computedNext, a rw2 model is fitted:
airp.rw2 <- inla(log(AirPassengers) ~ 0 + year + f(ID, model = "rw2"),
control.family = list(hyper = list(prec = list(param = c(1, 0.2)))),
data = airp.data, control.predictor = list(compute = TRUE))
summary(airp.rw2)##
## Call:
## c("inla(formula = log(AirPassengers) ~ 0 + year + f(ID, model =
## \"rw2\"), ", " data = airp.data, control.predictor = list(compute
## = TRUE), ", " control.family = list(hyper = list(prec = list(param
## = c(1, ", " 0.2)))))")
## Time used:
## Pre = 0.314, Running = 0.579, Post = 0.0333, Total = 0.927
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## year1949 5.148 0.278 4.602 5.148 5.694 5.148 0
## year1950 5.203 0.243 4.726 5.203 5.679 5.203 0
## year1951 5.323 0.210 4.909 5.323 5.736 5.323 0
## year1952 5.417 0.183 5.058 5.417 5.775 5.417 0
## year1953 5.467 0.162 5.149 5.467 5.784 5.467 0
## year1954 5.501 0.150 5.206 5.501 5.795 5.501 0
## year1955 5.585 0.149 5.291 5.585 5.879 5.585 0
## year1956 5.653 0.161 5.336 5.653 5.969 5.653 0
## year1957 5.715 0.182 5.357 5.715 6.073 5.715 0
## year1958 5.755 0.210 5.341 5.755 6.168 5.755 0
## year1959 5.847 0.243 5.369 5.847 6.325 5.847 0
## year1960 5.893 0.279 5.344 5.893 6.442 5.893 0
##
## Random effects:
## Name Model
## ID RW2 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 101.56 16.59 72.31 100.45
## Precision for ID 409.79 138.21 208.67 386.27
## 0.975quant mode
## Precision for the Gaussian observations 137.37 98.45
## Precision for ID 745.52 343.97
##
## Expected number of effective parameters(stdev): 47.16(4.53)
## Number of equivalent replicates : 3.05
##
## Marginal log-Likelihood: -12.71
## Posterior marginals for the linear predictor and
## the fitted values are computedNote that control.family has been used to set a prior on the precision of the
Gaussian likelihood to prevent overfitting (J. J. Faraway 2019b; Bakka 2019). See
also Chapter 5 on priors and Section 6.5.2.
In order to compare how these two models fit the data, the times series (in the
log scale) and the fitted values from these two models have been plotted in
Figure 3.3. In general, fitted values from both models are very
close to the observed time series. However, the estimates obtained with
the rw2 model seem to be smoother than those obtained with the rw1.
This is not surprising as the rw2 is of a higher order.
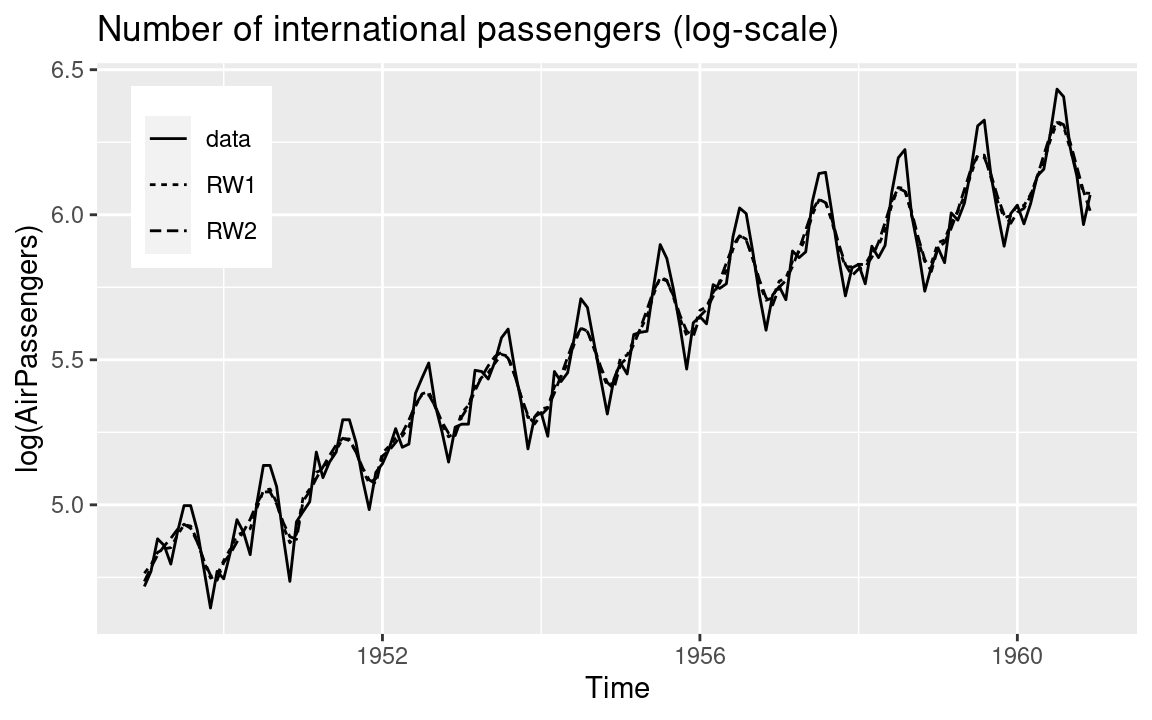
Figure 3.3: Fitted values (i.e., posterior means) of the number of international passengers.
Note that in this example, the latent effects are based on modeling the time series using values that are close in time, such as the one or two previous values. However, it is clear from Figure 3.2 that the time series has a strong season effect of period equal to one year. For this reason, it may be better to include a seasonal latent effect in the linear predictor.
3.3.10 Seasonal random effects
Seasonal effects can be modeled with the seasonal latent effect. In this
case, the vector of random effects \(\mathbf{u} = (u_1,\ldots, u_n)\) is assumed
to have periodicity \(p\) (with \(p < n\)) and it is defined such as sums \(x_i + x_{i+1} + \ldots + x_{i+m-1}\) are independent Gaussian observations with zero
mean and precision \(\tau\).
Note that the precision matrix of the model is \(Q = \tau R\), where \(R\) has the
neighborhood structure of the model. This model can be scaled, similarly to the
random walk models. See Table 3.8 for the different
specific options of this model. Note how periodicity of the seasonal
effect can be set with argument season.length when the model is defined
within the f() function.
For seasonal latent effects, the internal parameter is \(\theta = \log(\tau)\), and the default prior is a log-Gamma with parameters 1 and \(0.00005\). This prior can be modified as usual. See Section 3.5.1 and Chapter 5.
A seasonal model can be fit to the air passengers dataset as seen below.
Note that we have set the periodicity to 12 so that the latent effect
assumes that the seasonal effect covers one year.
This is reasonable because the time series shows a clear seasonal effect
of period equal to one year.
| Argument | Default | Description |
|---|---|---|
season.length |
— | Periodicity of the seasonal effect. |
scale.model |
FALSE |
Whether to scale variance matrix. |
Hence, the model is fit and summarized as follows:
airp.seasonal <- inla(log(AirPassengers) ~ 0 + year +
f(ID, model = "seasonal", season.length = 12),
control.family = list(hyper = list(prec = list(param = c(1, 0.2)))),
data = airp.data)
summary(airp.seasonal)##
## Call:
## c("inla(formula = log(AirPassengers) ~ 0 + year + f(ID, model =
## \"seasonal\", ", " season.length = 12), data = airp.data,
## control.family = list(hyper = list(prec = list(param = c(1, ", "
## 0.2)))))")
## Time used:
## Pre = 0.321, Running = 0.383, Post = 0.0281, Total = 0.732
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## year1949 4.836 0.02 4.797 4.836 4.875 4.836 0
## year1950 4.931 0.02 4.891 4.931 4.970 4.931 0
## year1951 5.131 0.02 5.092 5.131 5.171 5.131 0
## year1952 5.277 0.02 5.238 5.277 5.317 5.277 0
## year1953 5.409 0.02 5.369 5.409 5.448 5.409 0
## year1954 5.467 0.02 5.427 5.467 5.506 5.467 0
## year1955 5.639 0.02 5.600 5.639 5.679 5.639 0
## year1956 5.784 0.02 5.745 5.784 5.824 5.784 0
## year1957 5.898 0.02 5.859 5.898 5.938 5.898 0
## year1958 5.931 0.02 5.891 5.931 5.970 5.931 0
## year1959 6.048 0.02 6.009 6.048 6.088 6.048 0
## year1960 6.154 0.02 6.115 6.154 6.194 6.154 0
##
## Random effects:
## Name Model
## ID Seasonal model
##
## Model hyperparameters:
## mean sd 0.025quant
## Precision for the Gaussian observations 212.37 27.41 162.69
## Precision for ID 40021.83 24480.36 10396.94
## 0.5quant 0.975quant mode
## Precision for the Gaussian observations 211.00 270.28 208.66
## Precision for ID 34433.18 103017.17 24596.27
##
## Expected number of effective parameters(stdev): 26.52(1.85)
## Number of equivalent replicates : 5.43
##
## Marginal log-Likelihood: 70.223.3.11 Autoregressive random effects
Autoregressive models of order \(p\), denoted by AR(\(p\)), are also available in
INLA. In particular, an autoregressive model of order 1 (ar1) and a generic
order \(p\) (ar) are implemented.
The AR(1) model on a vector \(\mathbf{u}\) can be defined as \(u_1\) coming from a Gaussian distribution with zero mean and precision \(\tau (1 -\rho^2)\), where \(\tau\) is a precision parameter and \(\rho\) a correlation coefficient so that \(|\rho| < 1\). The other elements in \(\mathbf{u}\) are defined as \(u_i = \rho x_{i-1} + \varepsilon_i, i=2,\ldots,n\) with \(\varepsilon\) Gaussian observation with zero mean and precision \(\tau\).
Similarly as for the random walk models, autoregressive models can
take the values argument when defined to set the values of the
covariates for which the effects are being estimated. For the
ar1 model, covariates can be included using argument control.ar1c
when defining the model. Table 3.9 summarizes the specific
options of the autoregressive latent effects.
| Argument | Default | Description |
|---|---|---|
values |
— | Values of the covariate for which the effect is estimated. |
order |
— | Order of autoregressive model (for ar only). |
control.ar1c |
— | List with two objects: Z and Q.beta. |
control.ar1c$Z |
— | Matrix of covariates of dimension \(n\times m\). |
control.ar1c$Q.beta |
— | \(m\times m\) precision matrix of Gaussian prior on the coefficients of the covariates. |
This model is parameterized using \(\bm\theta = (\theta_1, \theta_2)\). The first internal hyperparameter is defined as \(\theta_1 = \log(\kappa)\), where \(\kappa\) is the marginal precision:
\[ \kappa = \tau (1 - \rho^2) \]
The second hyperparameter is defined as
\[ \theta_2 = \log(\frac{1 + \rho}{1 - \rho}) \]
The prior is defined on \(\theta\). The default is a log-Gamma with parameters \(1\) and \(0.00005\) for \(\theta_1\) and a Gaussian with zero mean and precision 1 for \(\theta_2\).
The AR(1) model can be extended to include covariates as follows:
\[ u_i = \rho u_{i-1} + \sum_{j=1}^m \beta_j z_{i-1}^{(j)}+\varepsilon_i, i=2,\ldots,n \]
This model is known as ar1c. Here, \(m\) covariates are used in the model, and
\(z_{i}^{(j)}\) is the value of the covariate \(j\) of observation \(i\). The
covariates have associated coefficients \(\beta_j, j=1,\ldots,m\).
Coefficients are assigned a multivariate Gaussian prior with known precision
matrix. The matrix of covariates (by column) and the precision of the prior on
the coefficients are defined in the f() function via parameter
control.ar1c, and they are passed as a list with names Z and Q.beta,
respectively. See Table 3.9 for details.
The AR(p) model is defined in a similar way, but it can now include a lag of order \(p\) on the vector of random effects:
\[ u_i = \varphi_1 x_{i-1} + \ldots + \varphi_p u_{i-p} + \varepsilon_i, i = p,\ldots, n \]
Note that this model can handle values of \(p\) up to 10 (inclusive).
The internal parameterization of this model is more complex and
we refer the reader to the manual page, which can be accessed
with inla.doc("^ar$"). The argument of inla.doc is due to the fact
that this function used regular expressions to look up in the documentation
files.
To illustrate the use of autoregressive models with INLA we will fit
a number of models to the AirPassengers dataset. We will start with an
AR(1) model:
airp.ar1 <- inla(log(AirPassengers) ~ 0 + year + f(ID, model = "ar1"),
control.family = list(hyper = list(prec = list(param = c(1, 0.2)))),
data = airp.data, control.predictor = list(compute = TRUE))
summary(airp.ar1)##
## Call:
## c("inla(formula = log(AirPassengers) ~ 0 + year + f(ID, model =
## \"ar1\"), ", " data = airp.data, control.predictor = list(compute
## = TRUE), ", " control.family = list(hyper = list(prec = list(param
## = c(1, ", " 0.2)))))")
## Time used:
## Pre = 0.328, Running = 0.203, Post = 0.0352, Total = 0.566
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## year1949 4.843 0.084 4.679 4.842 5.016 4.840 0
## year1950 4.940 0.081 4.782 4.939 5.104 4.938 0
## year1951 5.129 0.080 4.969 5.129 5.289 5.129 0
## year1952 5.277 0.080 5.117 5.277 5.436 5.277 0
## year1953 5.403 0.080 5.242 5.403 5.562 5.403 0
## year1954 5.478 0.080 5.320 5.477 5.640 5.476 0
## year1955 5.638 0.080 5.478 5.638 5.797 5.638 0
## year1956 5.776 0.080 5.614 5.777 5.934 5.778 0
## year1957 5.888 0.081 5.725 5.889 6.045 5.890 0
## year1958 5.932 0.080 5.771 5.933 6.091 5.933 0
## year1959 6.046 0.081 5.883 6.047 6.205 6.048 0
## year1960 6.124 0.085 5.948 6.127 6.287 6.130 0
##
## Random effects:
## Name Model
## ID AR1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 102.202 18.159 70.452 100.929
## Precision for ID 63.951 21.353 29.925 61.540
## Rho for ID 0.752 0.074 0.591 0.758
## 0.975quant mode
## Precision for the Gaussian observations 141.470 98.625
## Precision for ID 112.634 56.536
## Rho for ID 0.879 0.769
##
## Expected number of effective parameters(stdev): 62.15(8.37)
## Number of equivalent replicates : 2.32
##
## Marginal log-Likelihood: 4.69
## Posterior marginals for the linear predictor and
## the fitted values are computedNext, an AR(3) is fitted:
airp.ar3 <- inla(log(AirPassengers) ~ 0 + year + f(ID, model = "ar",
order = 3),
control.family = list(hyper = list(prec = list(param = c(1, 0.2)))),
data = airp.data, control.predictor = list(compute = TRUE))
summary(airp.ar3)##
## Call:
## c("inla(formula = log(AirPassengers) ~ 0 + year + f(ID, model =
## \"ar\", ", " order = 3), data = airp.data, control.predictor =
## list(compute = TRUE), ", " control.family = list(hyper = list(prec
## = list(param = c(1, ", " 0.2)))))")
## Time used:
## Pre = 0.332, Running = 0.563, Post = 0.0471, Total = 0.942
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## year1949 4.835 0.027 4.782 4.835 4.888 4.835 0
## year1950 4.930 0.027 4.877 4.930 4.983 4.930 0
## year1951 5.131 0.027 5.078 5.131 5.184 5.131 0
## year1952 5.277 0.027 5.225 5.277 5.330 5.277 0
## year1953 5.408 0.027 5.355 5.408 5.460 5.408 0
## year1954 5.465 0.027 5.412 5.465 5.518 5.465 0
## year1955 5.639 0.027 5.586 5.639 5.691 5.639 0
## year1956 5.784 0.027 5.731 5.784 5.837 5.784 0
## year1957 5.898 0.027 5.845 5.898 5.950 5.898 0
## year1958 5.930 0.027 5.877 5.930 5.983 5.930 0
## year1959 6.048 0.027 5.995 6.048 6.101 6.048 0
## year1960 6.154 0.027 6.101 6.154 6.207 6.154 0
##
## Random effects:
## Name Model
## ID AR(p) model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 122.636 12.136 99.027 122.679
## Precision for ID 181.637 94.418 55.927 163.237
## PACF1 for ID 0.849 0.004 0.841 0.848
## PACF2 for ID -0.757 0.045 -0.845 -0.756
## PACF3 for ID -0.948 0.013 -0.967 -0.951
## 0.975quant mode
## Precision for the Gaussian observations 146.707 123.526
## Precision for ID 417.007 127.378
## PACF1 for ID 0.859 0.847
## PACF2 for ID -0.663 -0.750
## PACF3 for ID -0.914 -0.956
##
## Expected number of effective parameters(stdev): 20.51(1.46)
## Number of equivalent replicates : 7.02
##
## Marginal log-Likelihood: 39.40
## Posterior marginals for the linear predictor and
## the fitted values are computedIn previous models covariate year has been included as part of the
linear predictor. In the next model this covariate is included as part
of an AR(1) model with covariates (i.e., a ar1c model):
Z <- model.matrix (~ 0 + year, data = airp.data)
Q.beta <- Diagonal(12, 0.001)
airp.ar1c <- inla(log(AirPassengers) ~ 1 + f(ID, model = "ar1c",
args.ar1c = list(Z = Z, Q.beta = Q.beta)),
control.family = list(hyper = list(prec = list(param = c(1, 0.2)))),
data = airp.data, control.predictor = list(compute = TRUE))
summary(airp.ar1c)##
## Call:
## c("inla(formula = log(AirPassengers) ~ 1 + f(ID, model = \"ar1c\",
## ", " args.ar1c = list(Z = Z, Q.beta = Q.beta)), data = airp.data,
## ", " control.predictor = list(compute = TRUE), control.family =
## list(hyper = list(prec = list(param = c(1, ", " 0.2)))))")
## Time used:
## Pre = 0.214, Running = 0.266, Post = 0.0362, Total = 0.516
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 4.724 0.135 4.458 4.724 4.99 4.724 0
##
## Random effects:
## Name Model
## ID AR1C model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 99.123 18.856 66.588 97.652
## Precision for ID 119.087 36.651 64.630 113.167
## Rho for ID 0.522 0.089 0.331 0.529
## 0.975quant mode
## Precision for the Gaussian observations 140.425 94.936
## Precision for ID 206.937 102.315
## Rho for ID 0.677 0.544
##
## Expected number of effective parameters(stdev): 60.32(9.96)
## Number of equivalent replicates : 2.39
##
## Marginal log-Likelihood: 3.65
## Posterior marginals for the linear predictor and
## the fitted values are computedIn all three models, the autoregressive effect seems to capture the high temporal autocorrelation in the data given the estimates of the autocorrelation parameter. Figure 3.4 shows the original time series (in the log scale) together with the posterior means of the fitted values.
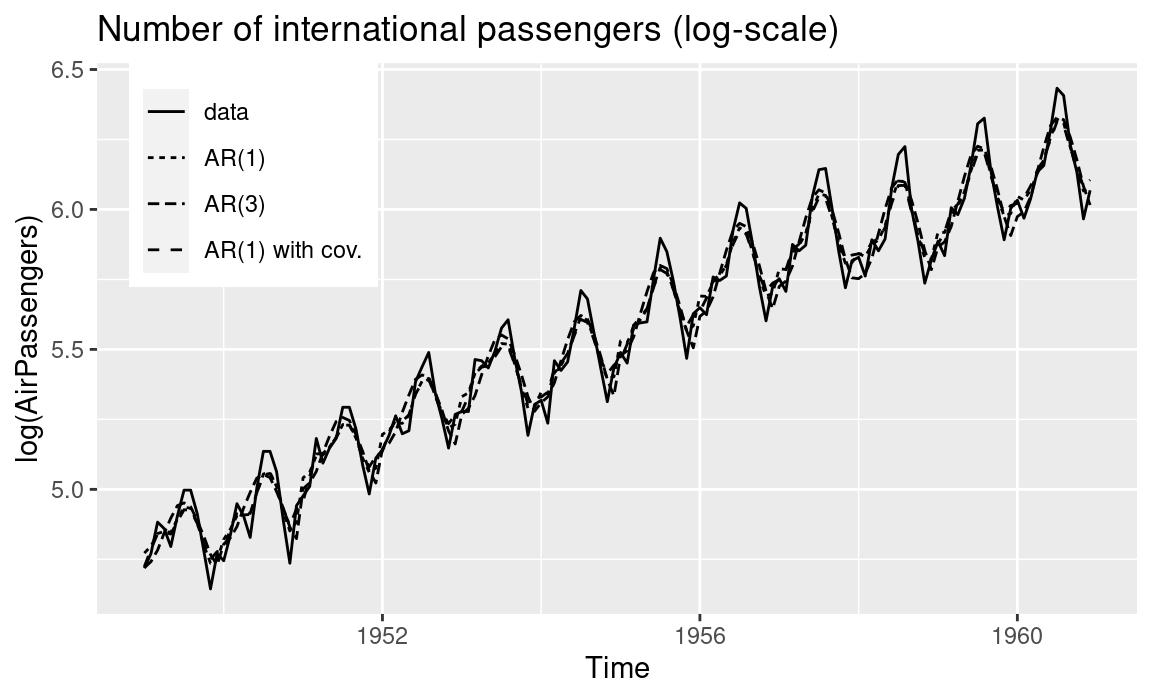
Figure 3.4: Fitted values (i.e., posterior means) of the number of international passengers using autoregressive models.
3.4 Information on the latent effects
The names of the hyperparameters in a given model, their internal
parameterization and the default prior on the internal scale can be checked in
the manual page (i.e., calling inla.doc()) or using function inla.models().
For example, the information about the idd model can be displayed with
inla.models()$latent$iid,
and similarly for any other random effects model.
This is particularly useful when in doubt about how a particular part of the model is defined, such as the internal representation of the parameters and their default priors. The full list of the elements that define the latent effect can be seen here:
names(inla.models()$latent$iid)## [1] "doc" "hyper" "constr"
## [4] "nrow.ncol" "augmented" "aug.factor"
## [7] "aug.constr" "n.div.by" "n.required"
## [10] "set.default.values" "pdf"Although particular random effects may require the use of specific parameters, there are a number of common parameters to all models that are worth knowing. These are described in the next section.
3.5 Additional arguments
In addition to the specific parameters required by each one of the
random effects previously described, there are a number of other arguments
that can be used to tune specific parts of the model. Some of these
are summarized in Table 3.10. All of them
must be passed to the f() function when defining the latent effect in the
model formula.
| Argument | Default | Description |
|---|---|---|
hyper |
Default priors | Definition of priors for hyperparameters. |
constr |
FALSE |
Sum-to-zero constraint on the random effects. |
extraconstr |
FALSE |
Additional constraint on the random effects. |
scale.model |
FALSE |
Scale variance matrix of random effects. |
group |
— | Variable to define the groups of random effect. |
control.group |
— | Control options for group parameter. |
3.5.1 Priors of hyperparameters
Priors in INLA are described in detail in Chapter 5 but we
include here a short introduction to how priors are set. Priors on the model
hyperparameters are set with parameter hyper, which takes a named list using
the names of the hyperparameters. Each element in the named list must be one of
the variables shown in Table 3.11.
For example, the default parameters for the prior on the log-precision of the
random effects of a iid random effects are:
prec.prior <- inla.models()$latent$iid$hyper$theta
prec.prior## $hyperid
## [1] 1001
## attr(,"inla.read.only")
## [1] FALSE
##
## $name
## [1] "log precision"
## attr(,"inla.read.only")
## [1] FALSE
##
## $short.name
## [1] "prec"
## attr(,"inla.read.only")
## [1] FALSE
##
## $prior
## [1] "loggamma"
## attr(,"inla.read.only")
## [1] FALSE
##
## $param
## [1] 1e+00 5e-05
## attr(,"inla.read.only")
## [1] FALSE
##
## $initial
## [1] 4
## attr(,"inla.read.only")
## [1] FALSE
##
## $fixed
## [1] FALSE
## attr(,"inla.read.only")
## [1] FALSE
##
## $to.theta
## function (x)
## log(x)
## <bytecode: 0x55a61a4dc528>
## <environment: 0x55a61a4d4c60>
## attr(,"inla.read.only")
## [1] TRUE
##
## $from.theta
## function (x)
## exp(x)
## <bytecode: 0x55a61a4dc640>
## <environment: 0x55a61a4d4c60>
## attr(,"inla.read.only")
## [1] TRUENote that in this case there is a single hyperparameter \(\theta\) and hence the
$theta after hyper. Other random effects with more than one hyperparameter
will name them theta1, etc.
In this example, the internal parameterization is \(\theta = \log(\tau)\)
(prec.prior$to.theta above), with \(\tau\) the precision of the iid random
effects. The default prior, on \(\theta\), is a log-Gamma (prec.prior$prior)
with parameters \(1\) and \(0.00005\) (prec.prior$param), as seen in
parameters prior and param. The starting value of \(\theta\) is \(4\), as seen
in parameter prec.prior$initial and it is not fixed by default (i.e., a posterior
marginal will be estimated) because prec.prior$fixed is FALSE. When fixed is set to
TRUE, the hyperparameter will be kept constant at the value in initial. In
this particular case, as initial is 4 this will make the log-precision to be
\(4\), i.e., \(4 = \theta = \log(\tau)\), which implies that the precision would be
fixed to \(\tau = \exp(4)\). This will represent a very large precision or,
equivalently, a very small variance which will make the random effects
very close to zero.
Furthermore, functions set in to.theta and from.theta are used to
convert the values of \(\tau\) to and from \(\theta\) (i.e., from the model scale
to and from the internal scale). In this case, given that \(\theta = \log(\tau)\), to.theta is the log() function while from.theta is the
exp() function, because \(\tau = \exp(\theta)\).
| Argument | Description |
|---|---|
hyperid |
Internal id of prior. |
name |
Name of hyperparameter in internal scale. |
short.name |
Short name of hyperparameter to be used inside hyper. |
prior |
Name of prior distribution. |
param |
Parameter values of prior distributions. |
initial |
Initial value of hyperparameter in the internal scale. |
fixed |
Whether to keep the hyperparameter fixed (default is FALSE). |
to.theta |
Function to convert parameter into the internal scale. |
from.theta |
Function to convert the value in the internal scale to the model scale. |
In general, not all arguments in the prior definition will be given
a value when setting a non-default prior. For
example, in the iid model the prior on the log-precision could be replaced by a
Gaussian with zero mean and small precision. This could be easily done
as follows:
hyper = list(prec = list(prior = "gaussian", param = c(0, 0.001)))The former R code sets a Gaussian prior with zero mean and precision
\(0.001\) on \(\theta\), i.e., a log-Gaussian prior on \(\tau\).
A full example on the North Carolina SIDS data using this prior would be:
m.pois.iid.gp <- inla(SID74 ~ f(ID, model = "iid",
hyper = list(theta = list(prior = "gaussian", param = c(0, 0.001)))),
data = nc.sids, family = "poisson",
E = EXP74)
summary(m.pois.iid.gp)##
## Call:
## c("inla(formula = SID74 ~ f(ID, model = \"iid\", hyper =
## list(theta = list(prior = \"gaussian\", ", " param = c(0,
## 0.001)))), family = \"poisson\", data = nc.sids, ", " E = EXP74)")
## Time used:
## Pre = 0.218, Running = 0.103, Post = 0.0183, Total = 0.339
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.032 0.064 -0.162 -0.031 0.091 -0.028 0
##
## Random effects:
## Name Model
## ID IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Precision for ID 6.52 2.15 3.49 6.14 11.82 5.52
##
## Expected number of effective parameters(stdev): 42.43(6.26)
## Number of equivalent replicates : 2.36
##
## Marginal log-Likelihood: -241.89The estimates of the parameters are very similar to the previous examples but
not equal and this is due to the choice of the priors. Note that in the named
list passed to hyper we have used prec to set the prior for the
hyperparameter. An alternative name could have been theta. i.e.,
hyper = list(theta = list(prior = "gaussian", param = c(0, 0.001)))However, using prec makes the code easier to read and avoids possible errors
when setting the priors for models with several hyperparameters. In this case,
the list passed to hyper can have length higher than one. For example, for a
model with two hyperparameters this would be:
hyper = list(theta1 = list(...), theta2 = list(...))Detailed information on how priors can be set in INLA (including implementing
new priors and other features) is given in Chapter 5.
3.5.2 Sum-to-zero constraint
Some of the models described above are intrinsic, i.e., their precision matrices are singular. Hence, the distribution is improper and may lead to an improper posterior. In other words, the precision matrix is not full-rank and additional constraints on the random effects are often imposed (see also Section 6.3).
A common one is a sum-to-zero constraint which will make the values of the
random effects \(\mathbf{u} = (u_1,\ldots, u_n)\) to be such as \(\sum_{i=1}^n u_i =0\).
This can be achieved by setting parameter constr to TRUE when the
model is defined. This sum-to-zero constraint is added by default
to all intrinsic models implemented in INLA.
For example, for a rw1 model the constr parameter is set to TRUE by default:
inla.models()$latent$rw1$constr## [1] TRUEOn the other hand, iid latent effects are not intrinsic and the
parameter is set to FALSE by default:
inla.models()$latent$iid$constr## [1] FALSENote that setting constraints on the random effects will likely affect the estimates of other parameters in the model as these constraints will shift the estimates of the random effects.
In the previous example on the North Carolina SIDS data, the way to add
a sum-to-zero constraint on the iid random effects (using the default
prior) would be:
m.pois.iid.gp0 <- inla(SID74 ~ f(ID, model = "iid", constr = TRUE),
data = nc.sids, family = "poisson", E = EXP74)
summary(m.pois.iid.gp0)##
## Call:
## c("inla(formula = SID74 ~ f(ID, model = \"iid\", constr = TRUE),
## family = \"poisson\", ", " data = nc.sids, E = EXP74)")
## Time used:
## Pre = 0.213, Running = 0.15, Post = 0.0182, Total = 0.381
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.027 0.049 -0.126 -0.026 0.067 -0.025 0
##
## Random effects:
## Name Model
## ID IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Precision for ID 7.26 2.57 3.79 6.77 13.61 6.02
##
## Expected number of effective parameters(stdev): 40.44(6.52)
## Number of equivalent replicates : 2.47
##
## Marginal log-Likelihood: -245.54In this particular case, the parameter estimates are quite similar.
3.5.3 Additional constraints on the random effects
In addition to the sum-to-zero constraint there is also the possibility of
setting additional constraints on the random effects.
See Section 6.3 for a general discussion on how to set
linear constraints on the latent effects in INLA.
Linear constraints are set with argument
extraconstr. This will take a list with two elements, A and e, to define
the constraint \(A \mathbf{u} = e\). Hence, A must be a matrix with the same
number of columns as the length of the vector of random effects, and
e must be a vector of the appropriate length, i.e., with the same
number of rows as matrix A.
For example, a sum-to-zero constraint for a vector of random effects
of length n could be added as follows:
A <- matrix(1, ncol = n, nrow = 1)
e <- rep(0, 1)
inla( ... ~ ... + f(..., extraconstr = list(A, e), ...), ...)Hence, the previous example could be reproduced as follows using
extraconstr:
n <- nrow(nc.sids)
A <- matrix(1, ncol = n, nrow = 1)
e <- rep(0, 1)
m.pois.iid.extrac <- inla(SID74 ~
f(ID, model = "iid", extraconstr = list(A = A, e = e)),
data = nc.sids, family = "poisson", E = EXP74)
summary(m.pois.iid.extrac)##
## Call:
## c("inla(formula = SID74 ~ f(ID, model = \"iid\", extraconstr =
## list(A = A, ", " e = e)), family = \"poisson\", data = nc.sids, E
## = EXP74)" )
## Time used:
## Pre = 0.223, Running = 0.146, Post = 0.0189, Total = 0.388
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -0.027 0.049 -0.126 -0.026 0.067 -0.025 0
##
## Random effects:
## Name Model
## ID IID model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant 0.975quant mode
## Precision for ID 7.26 2.57 3.79 6.77 13.61 6.02
##
## Expected number of effective parameters(stdev): 40.44(6.52)
## Number of equivalent replicates : 2.47
##
## Marginal log-Likelihood: -245.54Note how the fit model is the same as the previous one.
The main purpose to add additional constraint is for identifiability purposes. See, for example, Rue and Held (2005) for more details on this topic. Goicoa et al. (2018) show how additional constraints on the random effect can change the model estimates in spatio-temporal disease mapping models. However, this behavior is likely to be reproduced by models where the random effects are highly constrained for identifiability reasons.
3.5.4 Model scaling
Another feature that is particularly interesting when dealing with intrinsic models is the possibility to scale the model to make the diagonal of the structure variance (i.e., excluding the precision parameter) of the random effects to be re-scaled so that the generalized variance is equal to 1. This is fully described in Sørbye and Rue (2014), with a more practical example available in Sørbye (2013). Scaling in the context of prior setting is discussed later in Section 5.6.
The generalized variance is the geometric mean of the diagonal values of the covariance matrix (see Sørbye 2013 for details). By setting this to one the precision parameter is re-scaled as well. As a result of this scaling, the precision parameters of different models will have a similar interpretation. Hence, imposing the same priors on them (e.g., the default prior) will make sense as they are in the same scale. Furthermore, scaled models will be less sensible to the scale of the data used to estimate the effects (as shown in Sørbye 2013).
Scaling is set with parameter scale.model, which is FALSE by default. The
rw1 model fit to the air passengers data can be modified to scale the latent
random effect:
airp.rw1.scale <- inla(log(AirPassengers) ~ 0 + year +
f(ID, model = "rw1", scale.model = TRUE),
control.family = list(hyper = list(prec = list(param = c(1, 0.2)))),
data = airp.data)
summary(airp.rw1.scale)##
## Call:
## c("inla(formula = log(AirPassengers) ~ 0 + year + f(ID, model =
## \"rw1\", ", " scale.model = TRUE), data = airp.data,
## control.family = list(hyper = list(prec = list(param = c(1, ", "
## 0.2)))))")
## Time used:
## Pre = 0.313, Running = 0.377, Post = 0.0284, Total = 0.718
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## year1949 5.157 0.252 4.664 5.156 5.654 5.154 0
## year1950 5.207 0.220 4.776 5.206 5.641 5.205 0
## year1951 5.332 0.190 4.959 5.332 5.707 5.331 0
## year1952 5.420 0.165 5.096 5.420 5.745 5.420 0
## year1953 5.490 0.146 5.203 5.490 5.776 5.490 0
## year1954 5.517 0.135 5.251 5.517 5.782 5.517 0
## year1955 5.602 0.135 5.335 5.602 5.867 5.603 0
## year1956 5.670 0.146 5.382 5.670 5.956 5.671 0
## year1957 5.727 0.165 5.401 5.728 6.051 5.729 0
## year1958 5.736 0.190 5.360 5.736 6.109 5.737 0
## year1959 5.806 0.219 5.374 5.807 6.237 5.807 0
## year1960 5.843 0.251 5.348 5.843 6.335 5.844 0
##
## Random effects:
## Name Model
## ID RW1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 100.81 18.10 69.21 99.54
## Precision for ID 7.25 1.96 4.31 6.93
## 0.975quant mode
## Precision for the Gaussian observations 139.99 97.24
## Precision for ID 11.94 6.34
##
## Expected number of effective parameters(stdev): 58.89(8.79)
## Number of equivalent replicates : 2.44
##
## Marginal log-Likelihood: -221.27Figure 3.5 shows the effect of scaling a rw1 model. The
point estimates of the random effects do not seem to change but the posterior
marginals of the precision are clearly in different scales. Note that
this also means that the linear predictor is not affected by re-scaling
and the fitted values will be the same. For the scaled
model, the posterior marginal show smaller values than the one obtained with
the non-scaled model.
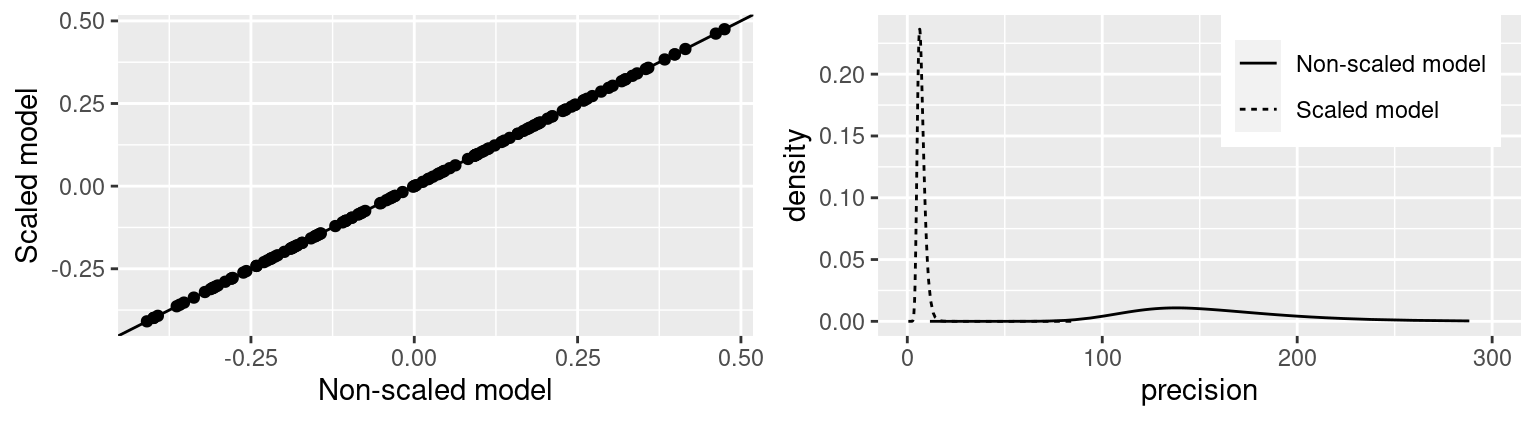
Figure 3.5: Non-scaled versus scaled models. Point estimates (i.e., posterior means) of the random effects (left) and posterior marginal of the precision of the random effect (right).
3.5.5 Grouped effects
Random effects can be grouped so that additional group-level correlation
can be included using an appropriate latent effect. The variable
that sets the group must be passed with parameter group and the actual model
and other parameters are passed through the control.group parameter.
This adds a new model to account for between group correlation, i.e.,
observations are already correlated using the primary random effects but also
according to the grouped effect. In practice, the variance structure of
the resulting model is the Kronecker product of the main and grouped random
effects. This is further discussed in Section 8.6, where
spatio-temporal models are discussed.
| Argument | Description |
|---|---|
adjust.for.con.comp |
Adjust for connected components when the model is scaled (default is TRUE). |
cyclic |
Make the group model cyclic (only for ar1, rw1 and rw2 models) |
graph |
Graph specification (for besag model). |
hyper |
Definition of priors in the hyperparameters. |
model |
Group model (exchangable, exchangablepos, ar1, ar, rw1, rw2, besag or iid). |
order |
Order of the model (only available for ar models). |
scale.model |
Scale the grouped model (default is TRUE; only for rw1, rw2 or besag models). |
Table 3.12 shows a summary of the options that can be passed
to control.group. In general, these are the same that apply to the random
effect when it is used.
In the next example we will analyze the air passengers data by using iid
random effects on the year but correlated random effects using an ar1 model
for the months. This implies that the observations for the same month of
consecutive years are correlated. First of all, two integer indices from 1 to 12
are created for the month (particularly important as this is passed to group)
and year, and then the model is fitted:
airp.data$month.num <- as.numeric(airp.data$month)
airp.data$year.num <- as.numeric(airp.data$year)
airp.iid.ar1 <- inla(log(airp) ~ 0 + f(year.num, model = "iid",
group = month.num, control.group = list(model = "ar1",
scale.model = TRUE)),
data = airp.data)
summary(airp.iid.ar1)##
## Call:
## c("inla(formula = log(airp) ~ 0 + f(year.num, model = \"iid\",
## group = month.num, ", " control.group = list(model = \"ar1\",
## scale.model = TRUE)), ", " data = airp.data)")
## Time used:
## Pre = 0.215, Running = 0.195, Post = 0.0218, Total = 0.432
## Random effects:
## Name Model
## year.num IID model
##
## Model hyperparameters:
## mean sd 0.025quant
## Precision for the Gaussian observations 2.22e+04 2.05e+04 2476.378
## Precision for year.num 4.70e-02 1.60e-02 0.022
## GroupRho for year.num 1.00e+00 0.00e+00 0.999
## 0.5quant 0.975quant mode
## Precision for the Gaussian observations 1.65e+04 7.64e+04 6969.55
## Precision for year.num 4.40e-02 8.40e-02 0.04
## GroupRho for year.num 1.00e+00 1.00e+00 1.00
##
## Expected number of effective parameters(stdev): 142.40(1.67)
## Number of equivalent replicates : 1.01
##
## Marginal log-Likelihood: 43.34Figure 3.6 shows the posterior estimates of the grouped random effects. Each plot represents a given month, and the increasing trend is clearly visible. In the model summary this can be appreciated by looking at the high correlation of the grouped effect, which has a posterior mean of 0.9997. Note also how summer months, for example, have higher estimates of the number of passengers.
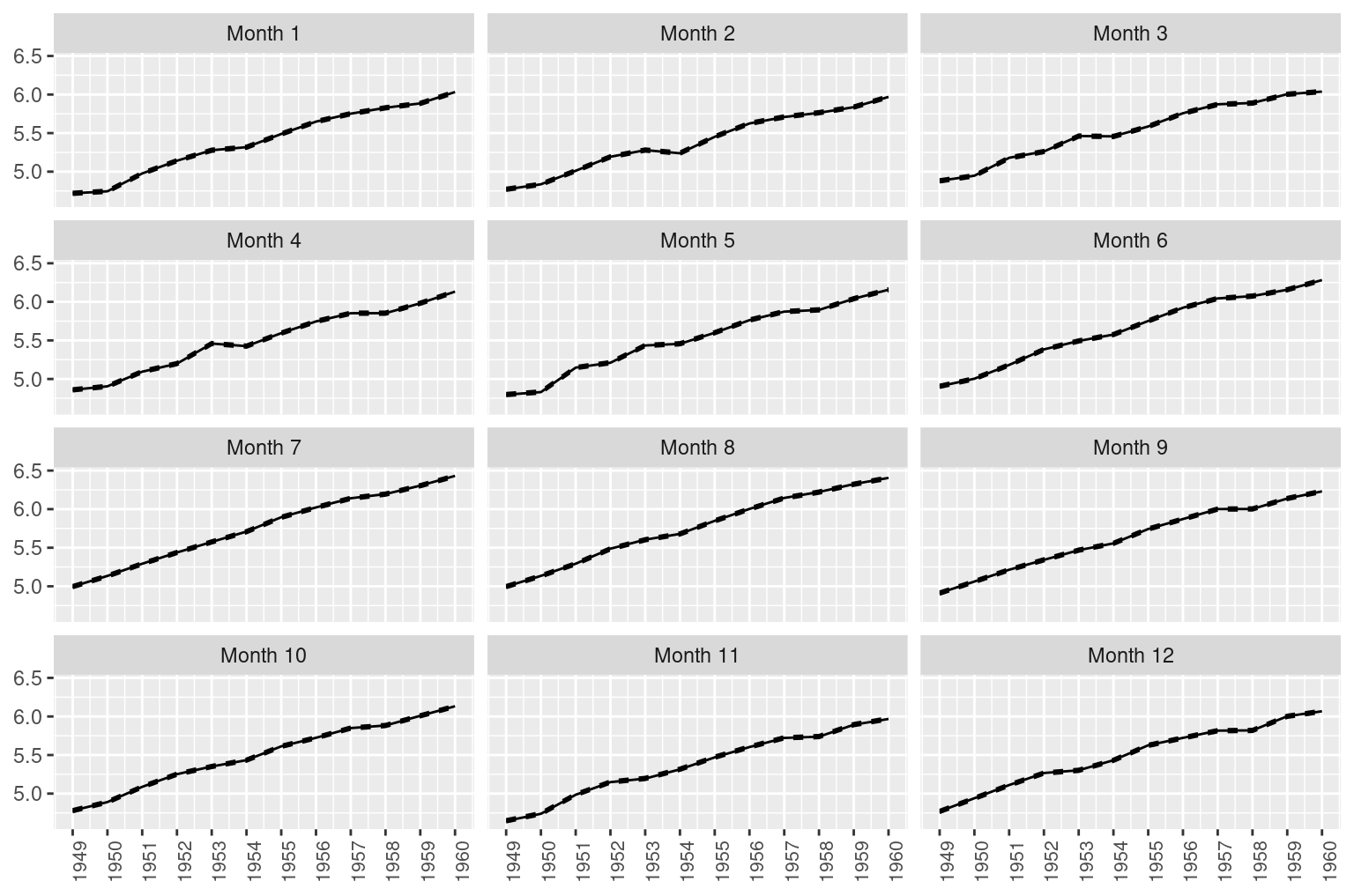
Figure 3.6: Grouped effects for the air passengers dataset. Solid lines represent the posterior means and the dashed lines 95% credible intervals.
3.6 Final remarks
In this chapter we have described some of the random effects implemented in
the INLA package. However, this list is far from exhaustive. Table 3.13 shows a list of other random effects not discussed in this chapter.
As INLA keeps growing, the list of available latent effects is likely
to increase too, so the list below may miss some recent developments.
Spatial random effects are described in detail in Chapter 7.
In general, detailed information about the random effects can be obtained from
the manual page that can be accessed with function inla.doc().
This function takes the name of the latent effect to be searched, e.g.,
inla.doc("ou").
| Model | Description |
|---|---|
ou |
Ornstein-Uhlenbeck process. |
sigm |
Sigmoidal effect of a covariate. |
revsigm |
Reverse-sigmoidal effect of a covariate. |
mec |
Classical measurement error model. |
meb |
Berkson Measurement Error model. |
matern2d |
Gaussian field with Matérn correlation in a 2D lattice. |
rw2d |
2-dimensional random walk. |
besag |
Besag spatial model. |
besag2 |
Weighted spatial effects model. |
besagproper |
Convolution spatial model. |
besagproper2 |
Convolution spatial model. |
bym |
Convolution spatial model. |
bym2 |
Reparameterized convolution spatial model. |
slm |
Spatial lag model. |
spde |
Spatial model based on Stochastic Partial Differential Equations. |
rgeneric |
Generic random effect. |
fgn |
Fractional Gaussian noise. |
fgn2 |
Reparameterized fractional Gaussian noise. |
copy |
Copy a random effect. |
log1exp |
Non-linear effect of a positive covariate. |
logdist |
Non-linear effect of a positive covariate. |
References
Bakka, Haakon. 2019. “Small Tutorials on INLA.” https://haakonbakka.bitbucket.io/organisedtopics.html.
Bivand, Roger S., Virgilio Gómez-Rubio, and HRue. 2014. “Approximate Bayesian Inference for Spatial Econometrics Models.” Spatial Statistics 9: 146–65.
Bivand, Roger S., Virgilio Gómez-Rubio, and Håvard Rue. 2015. “Spatial Data Analysis with R-INLA with Some Extensions.” Journal of Statistical Software 63 (20): 1–31. http://www.jstatsoft.org/v63/i20/.
Box, G. E. P., G. M. Jenkins, and G. C. Reinsel. 1976. Time Series Analysis, Forecasting and Control. 3rd ed. Holden-Day.
Faraway, Julian J. 2019b. “INLA for Linear Mixed Models.” http://www.maths.bath.ac.uk/~jjf23/inla/.
Goicoa, T., A. Adin, M. D. Ugarte, and J. S. Hodges. 2018. “In Spatio-Temporal Disease Mapping Models, Identifiability Constraints Affect PQL and INLA Results.” Stochastic Environmental Research and Risk Assessment 3 (32): 749–70.
Leroux, B., X. Lei, and N. Breslow. 1999. “Estimation of Disease Rates in Small Areas: A New Mixed Model for Spatial Dependence.” In Statistical Models in Epidemiology, the Environment and Clinical Trials, edited by M Halloran and D Berry, 135–78. New York: Springer-Verlag.
Pinheiro, José C., and Douglas M. Bates. 2000. Mixed-Effects Models in S and S-Plus. Springer, New York.
Rue, H., and L. Held. 2005. Gaussian Markov Random Fields: Theory and Applications. Chapman; Hall/CRC Press.
Sørbye, Sigrunn Holbek. 2013. Tutorial: Scaling Igmrf-Models in R-Inla.
Sørbye, Sigrunn Holbek, and Håvard Rue. 2014. “Scaling Intrinsic Gaussian Markov Random Field Priors in Spatial Modelling.” Spatial Statistics 8: 39–51. https://doi.org/https://doi.org/10.1016/j.spasta.2013.06.004.